The m6A reader YTHDC1 and the RNA helicase DDX5 control the production of rhabdomyosarcoma-enriched circRNAs
- PMID: 37019933
- PMCID: PMC10076346
- DOI: 10.1038/s41467-023-37578-7
The m6A reader YTHDC1 and the RNA helicase DDX5 control the production of rhabdomyosarcoma-enriched circRNAs
Abstract
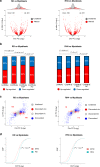
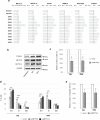
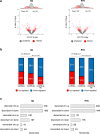
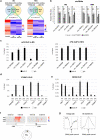
N6-Methyladenosine (m6A) is well-known for controlling different processes of linear RNA metabolism. Conversely, its role in the biogenesis and function of circular RNAs (circRNAs) is still poorly understood. Here, we characterize circRNA expression in the pathological context of rhabdomyosarcoma (RMS), observing a global increase when compared to wild-type myoblasts. For a set of circRNAs, such an increase is due to the raised expression of the m6A machinery, which we also find to control the proliferation activity of RMS cells. Furthermore, we identify the RNA helicase DDX5 as a mediator of the back-splicing reaction and as a co-factor of the m6A regulatory network. DDX5 and the m6A reader YTHDC1 are shown to interact and to promote the production of a common subset of circRNAs in RMS. In line with the observation that YTHDC1/DDX5 depletion reduces RMS proliferation, our results provide proteins and RNA candidates for the study of rhabdomyosarcoma tumorigenicity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Di Timoteo, G., Rossi, F. & Bozzoni, I. Circular RNAs in cell differentiation and development. Development147, dev182725. (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

