The KMT2A recombinome of acute leukemias in 2023
- PMID: 37019990
- PMCID: PMC10169636
- DOI: 10.1038/s41375-023-01877-1
The KMT2A recombinome of acute leukemias in 2023
Abstract
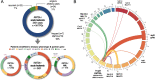
Chromosomal rearrangements of the human KMT2A/MLL gene are associated with de novo as well as therapy-induced infant, pediatric, and adult acute leukemias. Here, we present the data obtained from 3401 acute leukemia patients that have been analyzed between 2003 and 2022. Genomic breakpoints within the KMT2A gene and the involved translocation partner genes (TPGs) and KMT2A-partial tandem duplications (PTDs) were determined. Including the published data from the literature, a total of 107 in-frame KMT2A gene fusions have been identified so far. Further 16 rearrangements were out-of-frame fusions, 18 patients had no partner gene fused to 5'-KMT2A, two patients had a 5'-KMT2A deletion, and one ETV6::RUNX1 patient had an KMT2A insertion at the breakpoint. The seven most frequent TPGs and PTDs account for more than 90% of all recombinations of the KMT2A, 37 occur recurrently and 63 were identified so far only once. This study provides a comprehensive analysis of the KMT2A recombinome in acute leukemia patients. Besides the scientific gain of information, genomic breakpoint sequences of these patients were used to monitor minimal residual disease (MRD). Thus, this work may be directly translated from the bench to the bedside of patients and meet the clinical needs to improve patient survival.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

