Z-REX: shepherding reactive electrophiles to specific proteins expressed tissue specifically or ubiquitously, and recording the resultant functional electrophile-induced redox responses in larval fish
- PMID: 37020146
- PMCID: PMC11150335
- DOI: 10.1038/s41596-023-00809-8
Z-REX: shepherding reactive electrophiles to specific proteins expressed tissue specifically or ubiquitously, and recording the resultant functional electrophile-induced redox responses in larval fish
Erratum in
-
Author Correction: Z-REX: shepherding reactive electrophiles to specific proteins expressed tissue specifically or ubiquitously, and recording the resultant functional electrophile-induced redox responses in larval fish.Nat Protoc. 2023 Oct;18(10):3155. doi: 10.1038/s41596-023-00858-z. Nat Protoc. 2023. PMID: 37340167 No abstract available.
Abstract
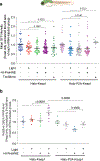
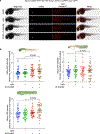
This Protocol Extension describes the adaptation of an existing Protocol detailing the use of targetable reactive electrophiles and oxidants, an on-demand redox targeting toolset in cultured cells. The adaptation described here is for use of reactive electrophiles and oxidants technologies in live zebrafish embryos (Z-REX). Zebrafish embryos expressing a Halo-tagged protein of interest (POI)-either ubiquitously or tissue specifically-are treated with a HaloTag-specific small-molecule probe housing a photocaged reactive electrophile (either natural electrophiles or synthetic electrophilic drug-like fragments). The reactive electrophile is then photouncaged at a user-defined time, enabling proximity-assisted electrophile-modification of the POI. Functional and phenotypic ramifications of POI-specific modification can then be monitored, by coupling to standard downstream assays, such as click chemistry-based POI-labeling and target-occupancy quantification; immunofluorescence or live imaging; RNA-sequencing and real-time quantitative polymerase chain reaction analyses of downstream-transcript modulations. Transient expression of requisite Halo-POI in zebrafish embryos is achieved by messenger RNA injection. Procedures associated with generation of transgenic zebrafish expressing a tissue-specific Halo-POI are also described. The Z-REX experiments can be completed in <1 week using standard techniques. To successfully execute Z-REX, researchers should have basic skills in fish husbandry, imaging and pathway analysis. Experience with protein or proteome manipulation is useful. This Protocol Extension is aimed at helping chemical biologists study precision redox events in a model organism and fish biologists perform redox chemical biology.
© 2023. Springer Nature Limited.
Figures













Similar articles
-
Monitoring On-Target Signaling Responses in Larval Zebrafish - Z-REX Unmasks Precise Mechanisms of Electrophilic Drugs and Metabolites.J Vis Exp. 2023 Jun 2;(196). doi: 10.3791/64846. J Vis Exp. 2023. PMID: 37335096
-
Single-Protein-Specific Redox Targeting in Live Mammalian Cells and C. elegans.Curr Protoc Chem Biol. 2018 Sep;10(3):e43. doi: 10.1002/cpch.43. Epub 2018 Aug 7. Curr Protoc Chem Biol. 2018. PMID: 30085412 Free PMC article.
-
T-REX on-demand redox targeting in live cells.Nat Protoc. 2016 Dec;11(12):2328-2356. doi: 10.1038/nprot.2016.114. Epub 2016 Oct 27. Nat Protoc. 2016. PMID: 27809314 Free PMC article.
-
Systems analysis of protein modification and cellular responses induced by electrophile stress.Acc Chem Res. 2010 May 18;43(5):673-83. doi: 10.1021/ar900286y. Acc Chem Res. 2010. PMID: 20218676 Free PMC article. Review.
-
Detection of electrophile-sensitive proteins.Biochim Biophys Acta. 2014 Feb;1840(2):913-22. doi: 10.1016/j.bbagen.2013.09.003. Epub 2013 Sep 8. Biochim Biophys Acta. 2014. PMID: 24021887 Free PMC article. Review.
Cited by
-
Toward decoding spatiotemporal signaling activities of reactive immunometabolites with precision immuno-chemical biology tools.Commun Chem. 2024 Sep 2;7(1):195. doi: 10.1038/s42004-024-01282-4. Commun Chem. 2024. PMID: 39223329 Free PMC article. Review.
References
-
- Lim W, Mayer B, Pawson T Cell Signaling: Principles and Mechanisms (Taylor & Francis, 2015).
-
- Murphy HC The use of whole animals versus isolated organs or cell culture in research. Trans. Nebr. Acad. Sci. Aff. Soc. XVIII, 105–108 (1991).
-
- Verdin E & Ott M 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015). - PubMed
-
- Komander D & Rape M The ubiquitin code. Ann. Rev. Biochem. 81, 203–229 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

