Efficient traceless modification of the P1 bacteriophage genome through homologous recombination with enrichment in double recombinants: A new perspective on the functional annotation of uncharacterized phage genes
- PMID: 37020717
- PMCID: PMC10067587
- DOI: 10.3389/fmicb.2023.1135870
Efficient traceless modification of the P1 bacteriophage genome through homologous recombination with enrichment in double recombinants: A new perspective on the functional annotation of uncharacterized phage genes
Abstract
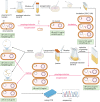
The advent of high-throughput omic technologies has caused unprecedented progress in research on bacteriophages, the most abundant and still the least explored entities on earth. Despite the growing number of phage genomes sequenced and the rejuvenation of interest in phage therapy, the progress in the functional analysis of phage genes is slow. Simple and efficient techniques of phage genome targeted mutagenesis that would allow one to knock out particular genes precisely without polar effects in order to study the effect of these knock-outs on phage functions are lacking. Even in the case of model phages, the functions of approximately half of their genes are unknown. P1 is an enterobacterial temperate myophage of clinical significance, which lysogenizes cells as a plasmid. It has a long history of studies, serves as a model in basic research, is a gene transfer vector, and is a source of genetic tools. Its gene products have structural homologs in several other phages. In this perspective article, we describe a simple and efficient procedure of traceless P1 genome modification that could also serve to acquire targeted mutations in the genomes of certain other temperate phages and speed up functional annotations of phage genes.
Keywords: ampicillin resistance; bacteriophage P1; bacteriophage engineering; homologous recombination; multicopy plasmid; prophage; targeted mutagenesis; temperate bacteriophage.
Copyright © 2023 Bednarek, Giermasińska-Buczek and Łobocka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Efficient Genome Engineering of a Virulent Klebsiella Bacteriophage Using CRISPR-Cas9.J Virol. 2018 Aug 16;92(17):e00534-18. doi: 10.1128/JVI.00534-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29899105 Free PMC article.
-
[Editing of Phage Genomes - Recombineering-Assisted SpCas9 Modification of Model Coliphages T7, T5, and T3].Mol Biol (Mosk). 2022 Nov-Dec;56(6):883. doi: 10.31857/S002689842206009X. Mol Biol (Mosk). 2022. PMID: 36475474 Russian.
-
Protein-Mediated and RNA-Based Origins of Replication of Extrachromosomal Mycobacterial Prophages.mBio. 2020 Mar 24;11(2):e00385-20. doi: 10.1128/mBio.00385-20. mBio. 2020. PMID: 32209683 Free PMC article.
-
Bacteriophage recombination systems and biotechnical applications.Appl Microbiol Biotechnol. 2014 Apr;98(7):2841-51. doi: 10.1007/s00253-014-5512-2. Epub 2014 Jan 19. Appl Microbiol Biotechnol. 2014. PMID: 24442504 Review.
-
Molecular ecology and evolution of Streptococcus thermophilus bacteriophages--a review.Virus Genes. 1998;16(1):95-109. doi: 10.1023/a:1007957911848. Virus Genes. 1998. PMID: 9562894 Review.
Cited by
-
The use of phages for the biosynthesis of metal nanoparticles and their biological applications: A review.J Cell Mol Med. 2024 Jun;28(11):e18383. doi: 10.1111/jcmm.18383. J Cell Mol Med. 2024. PMID: 38837580 Free PMC article. Review.
-
Interaction of bacteriophage P1 with an epiphytic Pantoea agglomerans strain-the role of the interplay between various mobilome elements.Front Microbiol. 2024 Mar 25;15:1356206. doi: 10.3389/fmicb.2024.1356206. eCollection 2024. Front Microbiol. 2024. PMID: 38591037 Free PMC article.
References
-
- Bai L., Wang J., Hurley D., Yu Z., Wang L., Chen Q., Li J., Li F., Fanning S. (2017). A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J. Antimicrob. Chemother. 72, 1531–1533. 10.1093/jac/dkw564 - DOI - PubMed
-
- Billard-Pomares T., Fouteau S., Jacquet M. E., Roche D., Barbe V., Castellanos M., et al. . (2014). Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 58, 6550–6557. 10.1128/AAC.03183-14 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

