Assessing arthropod diversity metrics derived from stream environmental DNA: spatiotemporal variation and paired comparisons with manual sampling
- PMID: 37020852
- PMCID: PMC10069422
- DOI: 10.7717/peerj.15163
Assessing arthropod diversity metrics derived from stream environmental DNA: spatiotemporal variation and paired comparisons with manual sampling
Abstract
Background: Benthic invertebrate (BI) surveys have been widely used to characterize freshwater environmental quality but can be challenging to implement at desired spatial scales and frequency. Environmental DNA (eDNA) allows an alternative BI survey approach, one that can potentially be implemented more rapidly and cheaply than traditional methods.
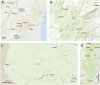
Methods: We evaluated eDNA analogs of BI metrics in the Potomac River watershed of the eastern United States. We first compared arthropod diversity detected with primers targeting mitochondrial 16S (mt16S) and cytochrome c oxidase 1 (cox1 or COI) loci to that detected by manual surveys conducted in parallel. We then evaluated spatial and temporal variation in arthropod diversity metrics with repeated sampling in three focal parks. We also investigated technical factors such as filter type used to capture eDNA and PCR inhibition treatment.
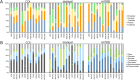
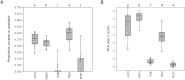
Results: Our results indicate that genus-level assessment of eDNA compositions is achievable at both loci with modest technical noise, although database gaps remain substantial at mt16S for regional taxa. While the specific taxa identified by eDNA did not strongly overlap with paired manual surveys, some metrics derived from eDNA compositions were rank-correlated with previously derived biological indices of environmental quality. Repeated sampling revealed statistical differences between high- and low-quality sites based on taxonomic diversity, functional diversity, and tolerance scores weighted by taxon proportions in transformed counts. We conclude that eDNA compositions are efficient and informative of stream condition. Further development and validation of scoring schemes analogous to commonly used biological indices should allow increased application of the approach to management needs.
Keywords: Arthropods; Benthic invertebrates; Biomonitoring; Freshwater streams; Metabarcoding; Water quality; eDNA.
© 2023 Aunins et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method.Sci Total Environ. 2022 Feb 1;806(Pt 2):150587. doi: 10.1016/j.scitotenv.2021.150587. Epub 2021 Sep 25. Sci Total Environ. 2022. PMID: 34582852
-
Taxonomic accuracy and complementarity between bulk and eDNA metabarcoding provides an alternative to morphology for biological assessment of freshwater macroinvertebrates.Sci Total Environ. 2024 Jul 20;935:173243. doi: 10.1016/j.scitotenv.2024.173243. Epub 2024 May 16. Sci Total Environ. 2024. PMID: 38761946
-
Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome c oxidase I primers.Mol Ecol Resour. 2018 Nov;18(6):1456-1468. doi: 10.1111/1755-0998.12940. Epub 2018 Sep 9. Mol Ecol Resour. 2018. PMID: 30129704
-
Effectiveness of eDNA for monitoring riverine macroinvertebrates.Sci Total Environ. 2024 Sep 1;941:173621. doi: 10.1016/j.scitotenv.2024.173621. Epub 2024 May 28. Sci Total Environ. 2024. PMID: 38815833 Review.
-
Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods.Sci Total Environ. 2022 Apr 20;818:151783. doi: 10.1016/j.scitotenv.2021.151783. Epub 2021 Nov 19. Sci Total Environ. 2022. PMID: 34801504 Review.
Cited by
-
Advancing the environmental DNA and RNA toolkit for aquatic ecosystem monitoring and management.PeerJ. 2025 Mar 18;13:e19119. doi: 10.7717/peerj.19119. eCollection 2025. PeerJ. 2025. PMID: 40124618 Free PMC article.
-
A Study on the Community and Ecological Characteristics of Benthic Invertebrates in the Ulungu River, Xinjiang, via eDNA Metabarcoding and Morphological Methods.Biology (Basel). 2025 Apr 12;14(4):410. doi: 10.3390/biology14040410. Biology (Basel). 2025. PMID: 40282275 Free PMC article.
References
-
- Alberdi A, Aizpurua O, Gilbert MTP, Bohmann K, Mahon A. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods in Ecology and Evolution. 2018;9(1):134–147. doi: 10.1111/2041-210X.12849. - DOI
-
- Allan JD. Landscapes and riverscapes: the influence of land use on stream ecosystems. Annual Review of Ecology, Evolution, and Systematics. 2004;35(1):257–284. doi: 10.1146/annurev.ecolsys.35.120202.110122. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

