Biocatalytic potential of Pseudolycoriella CAZymes (Sciaroidea, Diptera) in degrading plant and fungal cell wall polysaccharides
- PMID: 37020966
- PMCID: PMC10068558
- DOI: 10.1016/j.isci.2023.106449
Biocatalytic potential of Pseudolycoriella CAZymes (Sciaroidea, Diptera) in degrading plant and fungal cell wall polysaccharides
Abstract
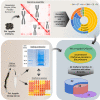
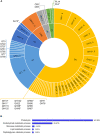
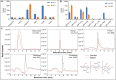
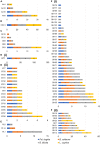
Soil biota has a crucial impact on soil ecology, global climate changes, and effective crop management and studying the diverse ecological roles of dipteran larvae deepens the understanding of soil food webs. A multi-omics study of Pseudolycoriella hygida comb. nov. (Diptera: Sciaroidea: Sciaridae) aimed to characterize carbohydrate-active enzymes (CAZymes) for litter degradation in this species. Manual curation of 17,881 predicted proteins in the Psl. hygida genome identified 137 secreted CAZymes, of which 33 are present in the saliva proteome, and broadly confirmed by saliva CAZyme catalytic profiling against plant cell wall polysaccharides and pNP-glycosyl substrates. Comparisons with two other sciarid species and the outgroup Lucilia cuprina (Diptera: Calliphoridae) identified 42 CAZyme families defining a sciarid CAZyme profile. The litter-degrading potential of sciarids corroborates their significant role as decomposers, yields insights to the evolution of insect feeding habits, and highlights the importance of insects as a source of biotechnologically relevant enzymes.
Keywords: Biocatalysis; Mycology; Plant biology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Lavelle P., Mathieu J., Spain A., Brown G., Fragoso C., Lapied E., De Aquino A., Barois I., Barrios E., Barros M.E., et al. Soil macroinvertebrate communities: a world-wide assessment. Global Ecol. Biogeogr. 2022;31:1261–1276. doi: 10.1111/geb.13492. - DOI
-
- Potapov A.M., Beaulieu F., Birkhofer K., Bluhm S.L., Degtyarev M.I., Devetter M., Goncharov A.A., Gongalsky K.B., Klarner B., Korobushkin D.I., et al. Feeding habits and multifunctional classification of soil-associated consumers from protists to vertebrates. Biol. Rev. Camb. Philos. Soc. 2022;97:1057–1117. doi: 10.1111/brv.12832. - DOI - PubMed
-
- Briones M.J.I. The serendipitous value of soil fauna in ecosystem functioning: the unexplained explained. Front. Environ. Sci. 2018;6 doi: 10.3389/fenvs.2018.00149. - DOI
-
- Guerra C.A., Heintz-Buschart A., Sikorski J., Chatzinotas A., Guerrero-Ramírez N., Cesarz S., Beaumelle L., Rillig M.C., Maestre F.T., Delgado-Baquerizo M., et al. Blind spots in global soil biodiversity and ecosystem function research. Nat. Commun. 2020;11:3870. doi: 10.1038/s41467-020-17688-2. - DOI - PMC - PubMed
-
- Gongalsky K.B. Soil macrofauna: study problems and perspectives. Soil Biol. Biochem. 2021;159 doi: 10.1016/j.soilbio.2021.108281. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

