First insight into the phylogeny of fine-leaved Festuca in the Altai Mountain Country based on genome-wide genotyping
- PMID: 37021080
- PMCID: PMC10067811
- DOI: 10.1002/ece3.9943
First insight into the phylogeny of fine-leaved Festuca in the Altai Mountain Country based on genome-wide genotyping
Abstract
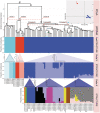
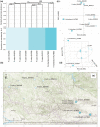
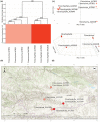
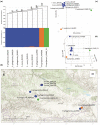
Festuca is one of the largest genera within the Poaceae family. Molecular phylogenies demonstrate that Festuca s.l. comprises two groups: broad- and fine-leaved species. The latter is the species-richest and taxonomically complicated group due to being paraphyletic. Here, we provide the first insight into the phylogeny of 17 fine-leaved species of Altai fescues. Based on genome-wide genotyping, the examined taxa were divided into three markedly differentiated clusters. The first cluster comprises species from the F. rubra complex, the second cluster includes the F. brachyphylla complex, and the third cluster contains taxa from the groups F. ovina, F. valesiaca, and F. kryloviana. Importantly, we detected a complex genetic pattern within the groups of F. valesiaca and F. kryloviana. Moreover, our findings underline a discrepancy between morphological and molecular data for some species distributed within the Altai Mountain Country. We suggest that in order to validate the current findings on the fine-leaved fescues, additional comprehensive research including morphological, karyological, and molecular methods is required. Nonetheless, our work provides a baseline for further investigations on the genus and studies on the floral diversity of Asia.
Keywords: Altai Mountain Country; DArTseq; distribution; fescues; hybridization; phylogeny.
© 2023 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Alexeev, E. B. (1976). New species of fescues (Festuca L.). Novosti Sistematiki Vysshikh Rastenii. Moscow, Leningrad, 13, 20–32.
-
- Alexeev, E. B. (1980). Ovsyanici Kavkaza. Izdatelstvo Moskovskogo Universiteta.
-
- Alexeev, E. B. (1984). On the new taxa and typification of some taxa of the genus Festuca (Poaceae) from South America. Botanicheskii Zhurnal, 69(3), 346–353.
-
- Alexeev, E. B. (1990). Festuca L. In Malyshev L. I. & Peschkova G. A. (Eds.), Flora Sibiriae Poaceae (Gramineae) (pp. 130–162). Nauka. Divisio Sibirica.
-
- Alexeev, E. B. , Sokolovskaya, A. P. , & Probatova, N. S. (1988). Taxonomy, distribution and chromosome numbers in Festuca L. (Gramineae) of the flora of USSR. 3. Section Festuca: F. tschujensis–F. beckeri. Bulleten Moskovskogo Obshchestva Ispytatelei Prirody . Otdel Biologicheskii, 93, 90–99.
Associated data
LinkOut - more resources
Full Text Sources

