Analysis of Donor Pancreata Defines the Transcriptomic Signature and Microenvironment of Early Neoplastic Lesions
- PMID: 37021392
- PMCID: PMC10236159
- DOI: 10.1158/2159-8290.CD-23-0013
Analysis of Donor Pancreata Defines the Transcriptomic Signature and Microenvironment of Early Neoplastic Lesions
Abstract
The adult healthy human pancreas has been poorly studied given the lack of indication to obtain tissue from the pancreas in the absence of disease and rapid postmortem degradation. We obtained pancreata from brain dead donors, thus avoiding any warm ischemia time. The 30 donors were diverse in age and race and had no known pancreas disease. Histopathologic analysis of the samples revealed pancreatic intraepithelial neoplasia (PanIN) lesions in most individuals irrespective of age. Using a combination of multiplex IHC, single-cell RNA sequencing, and spatial transcriptomics, we provide the first-ever characterization of the unique microenvironment of the adult human pancreas and of sporadic PanIN lesions. We compared healthy pancreata to pancreatic cancer and peritumoral tissue and observed distinct transcriptomic signatures in fibroblasts and, to a lesser extent, macrophages. PanIN epithelial cells from healthy pancreata were remarkably transcriptionally similar to cancer cells, suggesting that neoplastic pathways are initiated early in tumorigenesis.
Significance: Precursor lesions to pancreatic cancer are poorly characterized. We analyzed donor pancreata and discovered that precursor lesions are detected at a much higher rate than the incidence of pancreatic cancer, setting the stage for efforts to elucidate the microenvironmental and cell-intrinsic factors that restrain or, conversely, promote malignant progression. See related commentary by Hoffman and Dougan, p. 1288. This article is highlighted in the In This Issue feature, p. 1275.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures
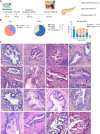
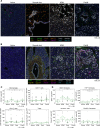

![Figure 4. Comparison of the microenvironment in healthy pancreata and pancreatic tumors reveals distinct stromal features. A, UMAP of all cells captured from single-cell RNA sequencing of six donor pancreata merged with 15 PDAC samples and 3 adjacent normal samples. Populations are identified by color. B, Histogram of cell-type abundance of all cell populations by disease state [healthy, adjacent normal (AdjNormal), and tumor]. C, Neighborhood graph differential abundance plot of the merged tumor, healthy, and adjacent normal samples. Size of dots represents neighborhoods, whereas edges represent the number of cells shared between neighborhoods. Neighborhoods colored in red represent significantly increased abundance in healthy samples, whereas neighborhoods colored in blue represent significantly increased abundance in tumor samples. logFC, log-fold change; Nhood, neighborhood. D, Beeswarm plot of differential abundance by cell type. X-axis represents a log-fold change in abundance between tumor and healthy states. Each dot is a neighborhood; neighborhoods colored in red represent significantly increased abundance in healthy samples, whereas neighborhoods colored in blue represent significantly increased abundance in tumor samples. E, Correlation heat map of pseudobulk-aggregrated counts of 15 tumor samples, 3 adjacent normal samples, and 11 donor samples (given that single-cell sequencing was performed on the head and tail sections separately in 5 out of 6 donors). Each row/line represents one aggregated single-cell sequencing sample. F, PCA plots of pseudobulk-aggregated counts from all cells, myeloid cells, T cells, and fibroblasts. Each dot represents one aggregated single-cell sequencing sample. G, Circos plots of putative ligand–receptor interactions that are upregulated in PDAC epithelial cells (left), fibroblasts (middle), and macrophages (right) compared with healthy cells. The heat map within the circos plots is the scaled average expression of each gene within PDAC tissue cell populations. The interactions plotted are those in which the expression level of the ligand is increased in PDAC samples compared with healthy tissues.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/92e4/10236159/19d1fe829f30/1324fig4.gif)



Update of
-
Analysis of donor pancreata defines the transcriptomic signature and microenvironment of early pre-neoplastic pancreatic lesions.bioRxiv [Preprint]. 2023 Jan 15:2023.01.13.523300. doi: 10.1101/2023.01.13.523300. bioRxiv. 2023. Update in: Cancer Discov. 2023 Jun 2;13(6):1324-1345. doi: 10.1158/2159-8290.CD-23-0013. PMID: 36712058 Free PMC article. Updated. Preprint.
Comment in
-
Pancreatic Neoplasms Are Frighteningly Common: A Role for Immune Surveillance?Cancer Discov. 2023 Jun 2;13(6):1288-1290. doi: 10.1158/2159-8290.CD-23-0332. Cancer Discov. 2023. PMID: 37264826
References
-
- Cocariu EA, Mageriu V, Staniceanu F, Bastian A, Socoliuc C, Zurac S. Correlations between the autolytic changes and postmortem interval in refrigerated cadavers. Rom J Intern Med 2016;54:105–12. - PubMed
-
- Shimizu M, Hayashi T, Saitoh Y, Ohta K, Itoh H. Postmortem autolysis in the pancreas: multivariate statistical study: the influence of clinicopathological conditions. Pancreas 1990;5:91–4. - PubMed
Publication types
MeSH terms
Grants and funding
- F31 CA265085/CA/NCI NIH HHS/United States
- P50 CA221707/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- I01 BX005777/BX/BLRD VA/United States
- R37 CA262209/CA/NCI NIH HHS/United States
- IK2 BX005875/BX/BLRD VA/United States
- U01 CA224145/CA/NCI NIH HHS/United States
- U54 CA274371/CA/NCI NIH HHS/United States
- T32 GM113900/GM/NIGMS NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- K08 CA234222/CA/NCI NIH HHS/United States
- R01 CA260752/CA/NCI NIH HHS/United States
- R37 CA214955/CA/NCI NIH HHS/United States
- R01 DK128102/DK/NIDDK NIH HHS/United States
- U01 CA274154/CA/NCI NIH HHS/United States
- R01 CA271510/CA/NCI NIH HHS/United States
- T32 GM141840/GM/NIGMS NIH HHS/United States
- T32 DK094775/DK/NIDDK NIH HHS/United States
- T32 AI007413/AI/NIAID NIH HHS/United States
- R50 CA232985/CA/NCI NIH HHS/United States
- R01 CA268426/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

