Chloroplastic Sec14-like proteins modulate growth and phosphate deficiency responses in Arabidopsis and rice
- PMID: 37021761
- PMCID: PMC10400038
- DOI: 10.1093/plphys/kiad212
Chloroplastic Sec14-like proteins modulate growth and phosphate deficiency responses in Arabidopsis and rice
Abstract
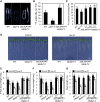
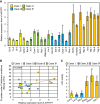
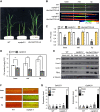
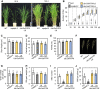
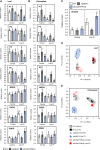
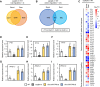
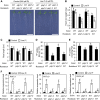
Phosphorus is an essential nutrient acquired from soil as phosphate (Pi), and its deficiency severely reduces plant growth and crop yield. Here, we show that single nucleotide polymorphisms (SNPs) at the PHOSPHATIDYLINOSITOL TRANSFER PROTEIN7 (AtPITP7) locus, which encodes a chloroplastic Sec14-like protein, are associated with genetic diversity regarding Pi uptake activity in Arabidopsis (Arabidopsis thaliana). Inactivation of AtPITP7 and its rice (Oryza sativa) homolog (OsPITP6) through T-DNA insertion and CRISPR/Cas9-mediated gene editing, respectively, decreased Pi uptake and plant growth, regardless of Pi availability. By contrast, overexpression of AtPITP7 and OsPITP6 enhanced Pi uptake and plant growth, especially under limited Pi supply. Importantly, overexpression of OsPITP6 increased the tiller number and grain yield in rice. Targeted metabolome analysis of glycerolipids in leaves and chloroplasts revealed that inactivation of OsPITP6 alters phospholipid contents, independent of Pi availability, diminishing the reduction in phospholipid content and increase in glycolipid content induced by Pi deficiency; meanwhile, overexpression of OsPITP6 enhanced Pi deficiency-induced metabolic alterations. Together with transcriptome analysis of ospitp6 rice plants and phenotypic analysis of grafted Arabidopsis chimeras, these results suggest that chloroplastic Sec14-like proteins play an essential role in growth modulations in response to changes in Pi availability, although their function is critical for plant growth under any Pi condition. The superior traits of OsPITP6-overexpressing rice plants also highlight the potential of OsPITP6 and its homologs in other crops as additional tools for improving Pi uptake and plant growth in low Pi environments.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

Comment in
-
Sec14-like proteins to the rescue: Improving plant performance in low-phosphate conditions.Plant Physiol. 2023 Aug 3;192(4):2588-2590. doi: 10.1093/plphys/kiad265. Plant Physiol. 2023. PMID: 37144824 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

