SpliceAI-10k calculator for the prediction of pseudoexonization, intron retention, and exon deletion
- PMID: 37021934
- PMCID: PMC10125908
- DOI: 10.1093/bioinformatics/btad179
SpliceAI-10k calculator for the prediction of pseudoexonization, intron retention, and exon deletion
Abstract
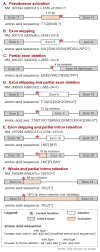
Summary: SpliceAI is a widely used splicing prediction tool and its most common application relies on the maximum delta score to assign variant impact on splicing. We developed the SpliceAI-10k calculator (SAI-10k-calc) to extend use of this tool to predict: the splicing aberration type including pseudoexonization, intron retention, partial exon deletion, and (multi)exon skipping using a 10 kb analysis window; the size of inserted or deleted sequence; the effect on reading frame; and the altered amino acid sequence. SAI-10k-calc has 95% sensitivity and 96% specificity for predicting variants that impact splicing, computed from a control dataset of 1212 single-nucleotide variants (SNVs) with curated splicing assay results. Notably, it has high performance (≥84% accuracy) for predicting pseudoexon and partial intron retention. The automated amino acid sequence prediction allows for efficient identification of variants that are expected to result in mRNA nonsense-mediated decay or translation of truncated proteins.
Availability and implementation: SAI-10k-calc is implemented in R (https://github.com/adavi4/SAI-10k-calc) and also available as a Microsoft Excel spreadsheet. Users can adjust the default thresholds to suit their target performance values.
© The Author(s) 2023. Published by Oxford University Press.
Figures
References
-
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF. et al. Predicting splicing from primary sequence with deep learning. Cell 2019;176:535–48.e24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

