SARS-CoV-2 Inhibits NRF2-Mediated Antioxidant Responses in Airway Epithelial Cells and in the Lung of a Murine Model of Infection
- PMID: 37022178
- PMCID: PMC10269779
- DOI: 10.1128/spectrum.00378-23
SARS-CoV-2 Inhibits NRF2-Mediated Antioxidant Responses in Airway Epithelial Cells and in the Lung of a Murine Model of Infection
Abstract
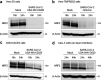
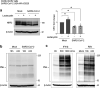
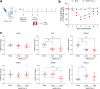
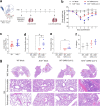
Several viruses have been shown to modulate the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), the master regulator of redox homeostasis. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the COVID-19 pandemic, also seems to disrupt the balance between oxidants and antioxidants, which likely contributes to lung damage. Using in vitro and in vivo models of infection, we investigated how SARS-CoV-2 modulates the transcription factor NRF2 and its dependent genes, as well as the role of NRF2 during SARS-CoV-2 infection. We found that SARS-CoV-2 infection downregulates NRF2 protein levels and NRF2-dependent gene expression in human airway epithelial cells and in lungs of BALB/c mice. Reductions in cellular levels of NRF2 seem to be independent of proteasomal degradation and the interferon/promyelocytic leukemia (IFN/PML) pathway. Furthermore, lack of the Nrf2 gene in SARS-CoV-2-infected mice exacerbates clinical disease, increases lung inflammation, and is associated with a trend toward increased lung viral titers, indicating that NRF2 has a protective role during this viral infection. In summary, our results suggest that SARS-CoV-2 infection alters the cellular redox balance by downregulating NRF2 and its dependent genes, which exacerbates lung inflammation and disease, therefore, suggesting that the activation of NRF2 could be explored as therapeutic approach during SARS-CoV-2 infection. IMPORTANCE The antioxidant defense system plays a major function in protecting the organism against oxidative damage caused by free radicals. COVID-19 patients often present with biochemical characteristics of uncontrolled pro-oxidative responses in the respiratory tract. We show herein that SARS-CoV-2 variants, including Omicron, are potent inhibitors of cellular and lung nuclear factor erythroid 2-related factor 2 (NRF2), the master transcription factor that controls the expression of antioxidant and cytoprotective enzymes. Moreover, we show that mice lacking the Nrf2 gene show increased clinical signs of disease and lung pathology when infected with a mouse-adapted strain of SARS-CoV-2. Overall, this study provides a mechanistic explanation for the observed unbalanced pro-oxidative response in SARS-CoV-2 infections and suggests that therapeutic strategies for COVID-19 may consider the use of pharmacologic agents that are known to boost the expression levels of cellular NRF2.
Keywords: COVID-19; NRF2; Nrf2-deficient mice; SARS-CoV-2; antioxidant enzymes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Dysregulation of intracellular redox homeostasis by the SARS-CoV-2 ORF6 protein.Virol J. 2023 Oct 18;20(1):239. doi: 10.1186/s12985-023-02208-7. Virol J. 2023. PMID: 37853388 Free PMC article.
-
Protective Role of Nuclear Factor Erythroid 2-Related Factor 2 Against Respiratory Syncytial Virus and Human Metapneumovirus Infections.Front Immunol. 2018 Apr 23;9:854. doi: 10.3389/fimmu.2018.00854. eCollection 2018. Front Immunol. 2018. PMID: 29740449 Free PMC article.
-
SARS-CoV-2 infection and dysregulation of nuclear factor erythroid-2-related factor 2 (Nrf2) pathway.Cell Stress Chaperones. 2023 Nov;28(6):657-673. doi: 10.1007/s12192-023-01379-0. Epub 2023 Oct 5. Cell Stress Chaperones. 2023. PMID: 37796433 Free PMC article. Review.
-
Cytokine storm in individuals with severe COVID-19 decreases endothelial cell antioxidant defense via downregulation of the Nrf2 transcriptional factor.Am J Physiol Heart Circ Physiol. 2023 Aug 1;325(2):H252-H263. doi: 10.1152/ajpheart.00096.2023. Epub 2023 Jun 16. Am J Physiol Heart Circ Physiol. 2023. PMID: 37327001 Free PMC article.
-
Transcription factor Nrf2 as a potential therapeutic target for COVID-19.Cell Stress Chaperones. 2023 Jan;28(1):11-20. doi: 10.1007/s12192-022-01296-8. Epub 2022 Nov 22. Cell Stress Chaperones. 2023. PMID: 36417098 Free PMC article. Review.
Cited by
-
Multiple Mechanisms of Action of Sulfodyne®, a Natural Antioxidant, against Pathogenic Effects of SARS-CoV-2 Infection.Antioxidants (Basel). 2024 Sep 4;13(9):1083. doi: 10.3390/antiox13091083. Antioxidants (Basel). 2024. PMID: 39334742 Free PMC article.
-
Dysregulation of intracellular redox homeostasis by the SARS-CoV-2 ORF6 protein.Virol J. 2023 Oct 18;20(1):239. doi: 10.1186/s12985-023-02208-7. Virol J. 2023. PMID: 37853388 Free PMC article.
-
The flavonoid quercetin decreases ACE2 and TMPRSS2 expression but not SARS-CoV-2 infection in cultured human lung cells.Biofactors. 2024 Nov-Dec;50(6):1268-1286. doi: 10.1002/biof.2084. Epub 2024 Jun 17. Biofactors. 2024. PMID: 38886986 Free PMC article.
-
Role of Hypoxia-Inducible Factors in Respiratory Syncytial Virus Infection-Associated Lung Disease.Int J Mol Sci. 2025 Mar 29;26(7):3182. doi: 10.3390/ijms26073182. Int J Mol Sci. 2025. PMID: 40244000 Free PMC article.
-
Metabolic Reprogramming in Respiratory Viral Infections: A Focus on SARS-CoV-2, Influenza, and Respiratory Syncytial Virus.Biomolecules. 2025 Jul 16;15(7):1027. doi: 10.3390/biom15071027. Biomolecules. 2025. PMID: 40723899 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

