Combined Methylation and Transcriptome Analysis of Liver Injury of Nonalcoholic Fatty Liver Disease Induced by High Alcohol-Producing Klebsiella pneumoniae
- PMID: 37022192
- PMCID: PMC10269619
- DOI: 10.1128/spectrum.05323-22
Combined Methylation and Transcriptome Analysis of Liver Injury of Nonalcoholic Fatty Liver Disease Induced by High Alcohol-Producing Klebsiella pneumoniae
Abstract
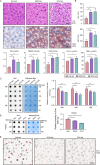
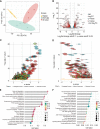
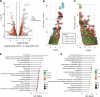
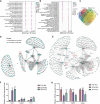
It has been known that high alcohol-producing Klebsiella pneumoniae (HiAlc Kpn) is one of causative agents of nonalcoholic fatty liver disease (NAFLD). However, how HiAlc Kpn promotes liver injury remains unclear. Recent findings suggest that DNA methylation might associate with the pathogenesis of NAFLD. Herein, the role of DNA methylation in HiAlc Kpn-induced liver injury was investigated. Murine models of NAFLD were established in C57BL/6N wild-type mice by gavaging HiAlc Kpn for 8 weeks. The liver injury was assessed based on the liver histopathology and biochemical indicators. In addition, DNA methylation in hepatic tissue was assessed by using dot bolt of 5-mC. RNA sequencing analysis and whole-genome bisulfite sequencing (WGBS) analysis were also performed. HiAlc Kpn significantly increased the activity of aspartate transaminase (AST), alanine transaminase (ALT), triglycerides (TGs), and glutathione (GSH), while hypomethylation was associated with liver injury in the experimental mice induced by HiAlc Kpn. The GO and KEGG pathway enrichment analysis of the transcriptome revealed that HiAlc Kpn induced fat metabolic disorders and DNA damage. The conjoint analysis of methylome and transcriptome showed that hypomethylation regulated related gene expression in signal pathways of lipid formation and circadian rhythm, including Rorα and Arntl1genes, which may be the dominant cause of NAFLD induced by HiAlc Kpn. Data suggest that DNA hypomethylation might play an important role in liver injury of NAFLD induced by HiAlc Kpn. Which possibly provides a new sight for understanding the mechanisms of NAFLD and selecting the potential therapeutic targets. IMPORTANCE High alcohol-producing Klebsiella pneumoniae (HiAlc Kpn) is one of causative agents of nonalcoholic fatty liver disease (NAFLD) and could induce liver damage. DNA methylation, as a common epigenetic form following contact with an etiologic agent and pathogenesis, can affect chromosome stability and transcription. We conjointly analyzed DNA methylation and transcriptome levels in the established murine models to explore the potential mechanisms for further understanding the role of DNA methylation in the liver damage of HiAlc Kpn-induced NAFLD. The analysis of the DNA methylation landscape contributes to our understanding of the entire disease process, which might be crucial in developing treatment strategies.
Keywords: DNA methylation; HiAlc Kpn; endogenous ethanol; liver injury.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, Cheng W, Li B, Li H, Lin W, Tian C, Zhao J, Han J, An D, Zhang Q, Wei H, Zheng M, Ma X, Li W, Chen X, Zhang Z, Zeng H, Ying S, Wu J, Yang R, Liu D. 2019. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab 30:1172. doi: 10.1016/j.cmet.2019.11.006. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

