A Longitudinal Study on the Dynamics of Salmonella enterica Prevalence and Serovar Composition in Beef Cattle Feces and Lymph Nodes and Potential Contributing Sources from the Feedlot Environment
- PMID: 37022263
- PMCID: PMC10132121
- DOI: 10.1128/aem.00033-23
A Longitudinal Study on the Dynamics of Salmonella enterica Prevalence and Serovar Composition in Beef Cattle Feces and Lymph Nodes and Potential Contributing Sources from the Feedlot Environment
Abstract
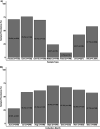
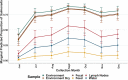
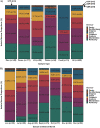
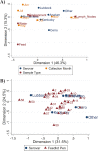
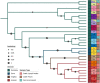
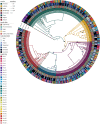
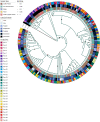
Salmonella can persist in the feedlot pen environment, acting as a source of transmission among beef cattle. Concurrently, cattle that are colonized with Salmonella can perpetuate contamination of the pen environment through fecal shedding. To study these cyclical dynamics, pen environment and bovine samples were collected for a 7-month longitudinal comparison of Salmonella prevalence, serovar, and antimicrobial resistance profiles. These samples included composite environment, water, and feed from the feedlot pens (n = 30) and cattle (n = 282) feces and subiliac lymph nodes. Salmonella prevalence across all sample types was 57.7%, with the highest prevalence in the pen environment (76.0%) and feces (70.9%). Salmonella was identified in 42.3% of the subiliac lymph nodes. Based on a multilevel mixed-effects logistic regression model, Salmonella prevalence varied significantly (P < 0.05) by collection month for most sample types. Eight Salmonella serovars were identified, and most isolates were pansusceptible, except for a point mutation in the parC gene, associated with fluoroquinolone resistance. There was a proportional difference in serovars Montevideo, Anatum, and Lubbock comparing the environment (37.2, 15.9, and 11.0%, respectively), fecal (27.5, 22.2, and 14.6%, respectively), and lymph node (15.6, 30.2, and 17.7%, respectively) samples. This suggests that the ability of Salmonella to migrate from the pen environment to the cattle host-or vice versa-is serovar specific. The presence of certain serovars also varied by season. Our results provide evidence that Salmonella serovar dynamics differ when comparing environment and host; therefore, developing serovar-specific preharvest environmental Salmonella mitigation strategies should be considered. IMPORTANCE Salmonella contamination of beef products, specifically from the incorporation of bovine lymph nodes into ground beef, remains a food safety concern. Current postharvest Salmonella mitigation techniques do not address Salmonella bacteria that are harbored in the lymph nodes, nor is it well understood how Salmonella invades the lymph nodes. Alternatively, preharvest mitigation techniques that can be applied to the feedlot environment, such as moisture applications, probiotics, or bacteriophage, may reduce Salmonella before dissemination into cattle lymph nodes. However, previous research conducted in cattle feedlots includes study designs that are cross-sectional, are limited to point-in-time sampling, or are limited to sampling of the cattle host, making it difficult to assess the Salmonella interactions between environment and hosts. This longitudinal analysis of the cattle feedlot explores the Salmonella dynamics between the feedlot environment and beef cattle over time to determine the applicability of preharvest environmental treatments.
Keywords: Salmonella enterica; feedlot cattle; feedlot pen environment; subiliac lymph nodes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA.
-
- Centers for Disease Control and Prevention. 2022. FoodNet FAST-Salmonella. Centers for Disease Control and Prevention, Atlanta, GA.
-
- Hanson D, Ison J, Malin K, Webb H. 2020. Salmonella white paper. Beef Industry Food Safety Council. https://www.bifsco.org/Media/BIFSCO/Docs/BIFSCo_SalmonellaWhitePaper_3-1....
-
- Le Minor L, Popoff M. 1987. Request for an opinion: designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Evol Microbiol 37:465–468.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

