Application of nanotags and nanobodies for live cell single-molecule imaging of the Z-ring in Escherichia coli
- PMID: 37022498
- PMCID: PMC10163087
- DOI: 10.1007/s00294-023-01266-2
Application of nanotags and nanobodies for live cell single-molecule imaging of the Z-ring in Escherichia coli
Abstract
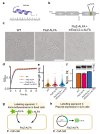
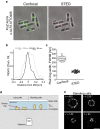
Understanding where proteins are localized in a bacterial cell is essential for understanding their function and regulation. This is particularly important for proteins that are involved in cell division, which localize at the division septum and assemble into highly regulated complexes. Current knowledge of these complexes has been greatly facilitated by super-resolution imaging using fluorescent protein fusions. Herein, we demonstrate with FtsZ that single-molecule PALM images can be obtained in-vivo using a genetically fused nanotag (ALFA), and a corresponding nanobody fused to mEos3.2. The methodology presented is applicable to other bacterial proteins.
Keywords: ALFA-tag; E. coli; FtsZ; Nanotags; PALM; STED.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

