Autopolyploidy, Allopolyploidy, and Phylogenetic Networks with Horizontal Arcs
- PMID: 37022524
- PMCID: PMC10079759
- DOI: 10.1007/s11538-023-01140-9
Autopolyploidy, Allopolyploidy, and Phylogenetic Networks with Horizontal Arcs
Abstract
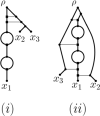
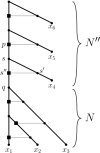
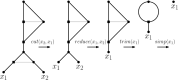
Polyploidization is an evolutionary process by which a species acquires multiple copies of its complete set of chromosomes. The reticulate nature of the signal left behind by it means that phylogenetic networks offer themselves as a framework to reconstruct the evolutionary past of species affected by it. The main strategy for doing this is to first construct a so-called multiple-labelled tree and to then somehow derive such a network from it. The following question therefore arises: How much can be said about that past if such a tree is not readily available? By viewing a polyploid dataset as a certain vector which we call a ploidy (level) profile, we show that among other results, there always exists a phylogenetic network in the form of a beaded phylogenetic tree with additional arcs that realizes a given ploidy profile. Intriguingly, the two end vertices of almost all of these additional arcs can be interpreted as having co-existed in time thereby adding biological realism to our network, a feature that is, in general, not enjoyed by phylogenetic networks. In addition, we show that our network may be viewed as a generator of ploidy profile space, a novel concept similar to phylogenetic tree space that we introduce to be able to compare phylogenetic networks that realize one and the same ploidy profile. We illustrate our findings in terms of a publicly available Viola dataset.
Keywords: Phylogenetic network; Ploidy profile; Ploidy profile space; Polyploid phylogenetics.
© 2023. The Author(s).
Figures
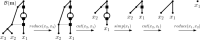
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

