High-Sensitivity Proteome-Scale Searches for Crosslinked Peptides Using CRIMP 2.0
- PMID: 37022750
- PMCID: PMC10353309
- DOI: 10.1021/acs.analchem.3c00329
High-Sensitivity Proteome-Scale Searches for Crosslinked Peptides Using CRIMP 2.0
Abstract
Crosslinking mass spectrometry (XL-MS) is a valuable technique for generating point-to-point distance measurements in protein space. However, cell-based XL-MS experiments require efficient software that can detect crosslinked peptides with sensitivity and controlled error rates. Many algorithms implement a filtering strategy designed to reduce the size of the database prior to mounting a search for crosslinks, but concern has been expressed over the possibility of reduced sensitivity using these strategies. We present a new scoring method that uses a rapid presearch method and a concept inspired by computer vision algorithms to resolve crosslinks from other conflicting reaction products. Searches of several curated crosslink datasets demonstrate high crosslink detection rates, and even the most complex proteome-level searches (using cleavable or noncleavable crosslinkers) can be completed efficiently on a conventional desktop computer. The detection of protein-protein interactions is increased twofold through the inclusion of compositional terms in the scoring equation. The combined functionality is made available as CRIMP 2.0 in the Mass Spec Studio.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



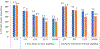

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

