Estimating the undetected emergence of COVID-19 in the US
- PMID: 37023065
- PMCID: PMC10079060
- DOI: 10.1371/journal.pone.0284025
Estimating the undetected emergence of COVID-19 in the US
Abstract
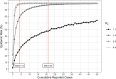
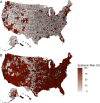
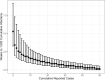
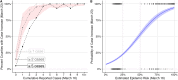
As SARS-CoV-2 emerged as a global threat in early 2020, China enacted rapid and strict lockdown orders to prevent introductions and suppress transmission. In contrast, the United States federal government did not enact national orders. State and local authorities were left to make rapid decisions based on limited case data and scientific information to protect their communities. To support local decision making in early 2020, we developed a model for estimating the probability of an undetected COVID-19 epidemic (epidemic risk) in each US county based on the epidemiological characteristics of the virus and the number of confirmed and suspected cases. As a retrospective analysis we included county-specific reproduction numbers and found that counties with only a single reported case by March 16, 2020 had a mean epidemic risk of 71% (95% CI: 52-83%), implying COVID-19 was already spreading widely by the first detected case. By that date, 15% of US counties covering 63% of the population had reported at least one case and had epidemic risk greater than 50%. We find that a 10% increase in model estimated epidemic risk for March 16 yields a 0.53 (95% CI: 0.49-0.58) increase in the log odds that the county reported at least two additional cases in the following week. The original epidemic risk estimates made on March 16, 2020 that assumed all counties had an effective reproduction number of 3.0 are highly correlated with our retrospective estimates (r = 0.99; p<0.001) but are less predictive of subsequent case increases (AIC difference of 93.3 and 100% weight in favor of the retrospective risk estimates). Given the low rates of testing and reporting early in the pandemic, taking action upon the detection of just one or a few cases may be prudent.
Copyright: © 2023 Javan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
SARS-CoV-2 pandemic and Construction Industry: insights from Italian data.Acta Biomed. 2022 Jul 1;93(3):e2022233. doi: 10.23750/abm.v93i3.12265. Acta Biomed. 2022. PMID: 35775779 Free PMC article.
-
Dynamic Panel Data Modeling and Surveillance of COVID-19 in Metropolitan Areas in the United States: Longitudinal Trend Analysis.J Med Internet Res. 2021 Feb 9;23(2):e26081. doi: 10.2196/26081. J Med Internet Res. 2021. PMID: 33481757 Free PMC article.
-
Association of Limited In-Person Attendance in US National Football League and National Collegiate Athletic Association Games With County-Level COVID-19 Cases.JAMA Netw Open. 2021 Aug 2;4(8):e2119621. doi: 10.1001/jamanetworkopen.2021.19621. JAMA Netw Open. 2021. PMID: 34402891 Free PMC article.
-
The early phase of the COVID-19 epidemic in Lombardy, Italy.Epidemics. 2021 Dec;37:100528. doi: 10.1016/j.epidem.2021.100528. Epub 2021 Nov 20. Epidemics. 2021. PMID: 34814093 Free PMC article.
-
Effect of national and local lockdowns on the control of COVID-19 pandemic: a rapid review.Epidemiol Prev. 2020 Sep-Dec;44(5-6 Suppl 2):60-68. doi: 10.19191/EP20.5-6.S2.104. Epidemiol Prev. 2020. PMID: 33412795 Review. English.
Cited by
-
An epidemic model for COVID-19 transmission in Argentina: Exploration of the alternating quarantine and massive testing strategies.Math Biosci. 2022 Apr;346:108664. doi: 10.1016/j.mbs.2021.108664. Epub 2021 Jul 13. Math Biosci. 2022. PMID: 34271015 Free PMC article.
References
-
- Ansari FM, Aggarwal K, Chopra A, Agrawal MG, Soni P, Agarwal P, et al.. Asymptomatic coronavirus: a boon or bane? J Adv Med Dent Sci Res. 2020;8: 109–111. doi: 10.21276/jamdsr - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

