African swine fever virus I73R is a critical virulence-related gene: A potential target for attenuation
- PMID: 37023125
- PMCID: PMC10104517
- DOI: 10.1073/pnas.2210808120
African swine fever virus I73R is a critical virulence-related gene: A potential target for attenuation
Abstract
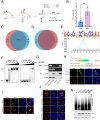
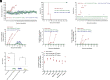
African swine fever virus (ASFV) is a large, double-stranded DNA virus that causes a fatal disease in pigs, posing a threat to the global pig industry. Whereas some ASFV proteins have been found to play important roles in ASFV-host interaction, the functional roles of many proteins are still largely unknown. In this study, we identified I73R, an early viral gene in the replication cycle of ASFV, as a key virulence factor. Our findings demonstrate that pI73R suppresses the host innate immune response by broadly inhibiting the synthesis of host proteins, including antiviral proteins. Crystallization and structural characterization results suggest that pI73R is a nucleic-acid-binding protein containing a Zα domain. It localizes in the nucleus and inhibits host protein synthesis by suppressing the nuclear export of cellular messenger RNA (mRNAs). While pI73R promotes viral replication, the deletion of the gene showed that it is a nonessential gene for virus replication. In vivo safety and immunogenicity evaluation results demonstrate that the deletion mutant ASFV-GZΔI73R is completely nonpathogenic and provides effective protection to pigs against wild-type ASFV. These results reveal I73R as a virulence-related gene critical for ASFV pathogenesis and suggest that it is a potential target for virus attenuation. Accordingly, the deletion mutant ASFV-GZΔI73R can be a potent live-attenuated vaccine candidate.
Keywords: ASFV; I73R; immune suppression; live-attenuated vaccine candidate; single-stranded RNA binding.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Alonso C., et al. , ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 99, 613–614 (2018). - PubMed
-
- Zhou X., et al. , Emergence of African Swine fever in China, 2018. Transbound. Emerg. Dis. 65, 1482–1484 (2018). - PubMed
-
- Tao D., et al. , One year of African swine fever outbreak in China. Acta Trop. 211, 105602 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

