Starships are active eukaryotic transposable elements mobilized by a new family of tyrosine recombinases
- PMID: 37023132
- PMCID: PMC10104507
- DOI: 10.1073/pnas.2214521120
Starships are active eukaryotic transposable elements mobilized by a new family of tyrosine recombinases
Abstract
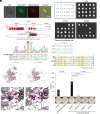
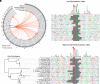
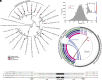
Transposable elements in eukaryotic organisms have historically been considered "selfish," at best conferring indirect benefits to their host organisms. The Starships are a recently discovered feature in fungal genomes that are, in some cases, predicted to confer beneficial traits to their hosts and also have hallmarks of being transposable elements. Here, we provide experimental evidence that Starships are indeed autonomous transposons, using the model Paecilomyces variotii, and identify the HhpA "Captain" tyrosine recombinase as essential for their mobilization into genomic sites with a specific target site consensus sequence. Furthermore, we identify multiple recent horizontal gene transfers of Starships, implying that they jump between species. Fungal genomes have mechanisms to defend against mobile elements, which are frequently detrimental to the host. We discover that Starships are also vulnerable to repeat-induced point mutation defense, thereby having implications on the evolutionary stability of such elements.
Keywords: Starship; fungi; horizontal gene transfer; transposon.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Starships: a new frontier for fungal biology.Trends Genet. 2024 Dec;40(12):1060-1073. doi: 10.1016/j.tig.2024.08.006. Epub 2024 Sep 18. Trends Genet. 2024. PMID: 39299886 Review.
-
That's no moon, it's a Starship: Giant transposons driving fungal horizontal gene transfer.Mol Microbiol. 2023 Oct;120(4):555-563. doi: 10.1111/mmi.15118. Epub 2023 Jul 11. Mol Microbiol. 2023. PMID: 37434470
-
Gene acquisition by giant transposons primes eukaryotes for rapid evolution via horizontal gene transfer.Sci Adv. 2024 Dec 6;10(49):eadp8738. doi: 10.1126/sciadv.adp8738. Epub 2024 Dec 6. Sci Adv. 2024. PMID: 39642232 Free PMC article.
-
Starship giant transposable elements cluster by host taxonomy using k-mer-based phylogenetics.G3 (Bethesda). 2025 Jun 4;15(6):jkaf082. doi: 10.1093/g3journal/jkaf082. G3 (Bethesda). 2025. PMID: 40211926 Free PMC article.
-
Horizontal transfers of transposable elements in eukaryotes: The flying genes.C R Biol. 2016 Jul-Aug;339(7-8):296-9. doi: 10.1016/j.crvi.2016.04.013. Epub 2016 May 24. C R Biol. 2016. PMID: 27234293 Review.
Cited by
-
Giant transposons promote strain heterogeneity in a major fungal pathogen.bioRxiv [Preprint]. 2025 Apr 6:2024.06.28.601215. doi: 10.1101/2024.06.28.601215. bioRxiv. 2025. Update in: mBio. 2025 Jun 11;16(6):e0109225. doi: 10.1128/mbio.01092-25. PMID: 38979181 Free PMC article. Updated. Preprint.
-
A new Paecilomyces from wooden utility poles in South Africa.Fungal Syst Evol. 2024 Jun;13:163-181. doi: 10.3114/fuse.2024.13.10. Epub 2024 Jun 7. Fungal Syst Evol. 2024. PMID: 39140099 Free PMC article.
-
Horizontal transfers between fungal Fusarium species contributed to successive outbreaks of coffee wilt disease.PLoS Biol. 2024 Dec 5;22(12):e3002480. doi: 10.1371/journal.pbio.3002480. eCollection 2024 Dec. PLoS Biol. 2024. PMID: 39637834 Free PMC article.
-
Systematic identification of cargo-mobilizing genetic elements reveals new dimensions of eukaryotic diversity.Nucleic Acids Res. 2024 Jun 10;52(10):5496-5513. doi: 10.1093/nar/gkae327. Nucleic Acids Res. 2024. PMID: 38686785 Free PMC article.
-
Gene transfer between fungal species triggers repeated coffee wilt disease outbreaks.PLoS Biol. 2024 Dec 6;22(12):e3002901. doi: 10.1371/journal.pbio.3002901. eCollection 2024 Dec. PLoS Biol. 2024. PMID: 39642107 Free PMC article.
References
-
- Cantrell S. A., Dianese J. C., Fell J., Gunde-Cimerman N., Zalar P., Unusual fungal niches. Mycologia 103, 1161–1174 (2011). - PubMed
-
- Stukenbrock E. H., Croll D., The evolving fungal genome. Fungal. Biol. Rev. 28, 1–12 (2014).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

