The defense island repertoire of the Escherichia coli pan-genome
- PMID: 37023146
- PMCID: PMC10121019
- DOI: 10.1371/journal.pgen.1010694
The defense island repertoire of the Escherichia coli pan-genome
Abstract
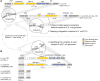
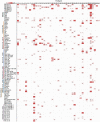
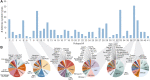
It has become clear in recent years that anti-phage defense systems cluster non-randomly within bacterial genomes in so-called "defense islands". Despite serving as a valuable tool for the discovery of novel defense systems, the nature and distribution of defense islands themselves remain poorly understood. In this study, we comprehensively mapped the defense system repertoire of >1,300 strains of Escherichia coli, the most widely studied organism for phage-bacteria interactions. We found that defense systems are usually carried on mobile genetic elements including prophages, integrative conjugative elements and transposons, which preferentially integrate at several dozens of dedicated hotspots in the E. coli genome. Each mobile genetic element type has a preferred integration position but can carry a diverse variety of defensive cargo. On average, an E. coli genome has 4.7 hotspots occupied by defense system-containing mobile elements, with some strains possessing up to eight defensively occupied hotspots. Defense systems frequently co-localize with other systems on the same mobile genetic element, in agreement with the observed defense island phenomenon. Our data show that the overwhelming majority of the E. coli pan-immune system is carried on mobile genetic elements, explaining why the immune repertoire varies substantially between different strains of the same species.
Copyright: © 2023 Hochhauser et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: R.S. is a scientific cofounder and advisor of BiomX and Ecophage. The other authors declare that they have no competing interests.
Figures



Similar articles
-
Exploring the diversity of anti-defense systems across prokaryotes, phages, and mobile genetic elements.bioRxiv [Preprint]. 2024 Aug 21:2024.08.21.608784. doi: 10.1101/2024.08.21.608784. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jan 07;53(1):gkae1171. doi: 10.1093/nar/gkae1171. PMID: 39229129 Free PMC article. Updated. Preprint.
-
Pipolins are bimodular platforms that maintain a reservoir of defense systems exchangeable with various bacterial genetic mobile elements.Nucleic Acids Res. 2024 Nov 11;52(20):12498-12516. doi: 10.1093/nar/gkae891. Nucleic Acids Res. 2024. PMID: 39404074 Free PMC article.
-
Phages and their satellites encode hotspots of antiviral systems.Cell Host Microbe. 2022 May 11;30(5):740-753.e5. doi: 10.1016/j.chom.2022.02.018. Epub 2022 Mar 21. Cell Host Microbe. 2022. PMID: 35316646 Free PMC article.
-
The molecular basis of infectious diseases: pathogenicity islands and other mobile genetic elements. A review.Acta Microbiol Immunol Hung. 2003;50(4):321-30. doi: 10.1556/AMicr.50.2003.4.1. Acta Microbiol Immunol Hung. 2003. PMID: 14750434 Review.
-
[Plasticity of bacterial genomes: pathogenicity islands and the locus of enterocyte effacement (LEE)].Berl Munch Tierarztl Wochenschr. 2004 Mar-Apr;117(3-4):116-29. Berl Munch Tierarztl Wochenschr. 2004. PMID: 15046458 Review. German.
Cited by
-
Mobile genetic elements: the hidden puppet masters underlying infant gut microbiome assembly?Microbiome Res Rep. 2024 Nov 9;4(1):7. doi: 10.20517/mrr.2024.51. eCollection 2025. Microbiome Res Rep. 2024. PMID: 40207272 Free PMC article.
-
Antiviral defence arsenal across members of the Bacillus cereus group.Sci Rep. 2025 Feb 10;15(1):4958. doi: 10.1038/s41598-025-86748-8. Sci Rep. 2025. PMID: 39929895 Free PMC article.
-
Pseudomonas aeruginosa as a model bacterium in antiphage defense research.FEMS Microbiol Rev. 2025 Jan 14;49:fuaf014. doi: 10.1093/femsre/fuaf014. FEMS Microbiol Rev. 2025. PMID: 40240293 Free PMC article. Review.
-
LoVis4u: a locus visualization tool for comparative genomics and coverage profiles.NAR Genom Bioinform. 2025 Feb 24;7(1):lqaf009. doi: 10.1093/nargab/lqaf009. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 40007724 Free PMC article.
-
Unveiling the multifaceted domain polymorphism of the Menshen antiphage system.Nucleic Acids Res. 2025 May 10;53(9):gkaf357. doi: 10.1093/nar/gkaf357. Nucleic Acids Res. 2025. PMID: 40347139 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

