Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
- PMID: 37023263
- PMCID: PMC10119980
- DOI: 10.1021/jacs.3c01003
Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
Abstract
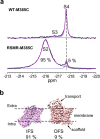
Limited chemical shift dispersion represents a significant barrier to studying multistate equilibria of large membrane proteins by 19F NMR. We describe a novel monofluoroethyl 19F probe that dramatically increases the chemical shift dispersion. The improved conformational sensitivity and line shape enable the detection of previously unresolved states in one-dimensional (1D) 19F NMR spectra of a 134 kDa membrane transporter. Changes in the populations of these states in response to ligand binding, mutations, and temperature correlate with population changes of distinct conformations in structural ensembles determined by single-particle cryo-electron microscopy (cryo-EM). Thus, 19F NMR can guide sample preparation to discover and visualize novel conformational states and facilitate image analysis and three-dimensional (3D) classification.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

