An OMA1 redox site controls mitochondrial homeostasis, sarcoma growth, and immunogenicity
- PMID: 37024121
- PMCID: PMC10078952
- DOI: 10.26508/lsa.202201767
An OMA1 redox site controls mitochondrial homeostasis, sarcoma growth, and immunogenicity
Abstract
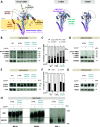
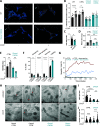
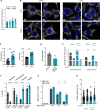
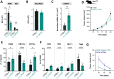
Aggressive tumors often display mitochondrial dysfunction. Upon oxidative stress, mitochondria undergo fission through OMA1-mediated cleavage of the fusion effector OPA1. In yeast, a redox-sensing switch participates in OMA1 activation. 3D modeling of OMA1 comforted the notion that cysteine 403 might participate in a similar sensor in mammalian cells. Using prime editing, we developed a mouse sarcoma cell line in which OMA1 cysteine 403 was mutated in alanine. Mutant cells showed impaired mitochondrial responses to stress including ATP production, reduced fission, resistance to apoptosis, and enhanced mitochondrial DNA release. This mutation prevented tumor development in immunocompetent, but not nude or cDC1 dendritic cell-deficient, mice. These cells prime CD8+ lymphocytes that accumulate in mutant tumors, whereas their depletion delays tumor control. Thus, OMA1 inactivation increased the development of anti-tumor immunity. Patients with complex genomic soft tissue sarcoma showed variations in the level of OMA1 and OPA1 transcripts. High expression of OPA1 in primary tumors was associated with shorter metastasis-free survival after surgery, and low expression of OPA1, with anti-tumor immune signatures. Targeting OMA1 activity may enhance sarcoma immunogenicity.
© 2023 Miallot et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
