Distributed processing for value-based choice by prelimbic circuits targeting anterior-posterior dorsal striatal subregions in male mice
- PMID: 37024449
- PMCID: PMC10079960
- DOI: 10.1038/s41467-023-36795-4
Distributed processing for value-based choice by prelimbic circuits targeting anterior-posterior dorsal striatal subregions in male mice
Abstract
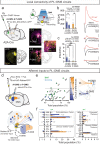
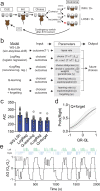
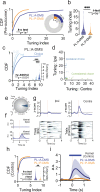
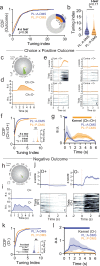
Fronto-striatal circuits have been implicated in cognitive control of behavioral output for social and appetitive rewards. The functional diversity of prefrontal cortical populations is strongly dependent on their synaptic targets, with control of motor output mediated by connectivity to dorsal striatum. Despite evidence for functional diversity along the anterior-posterior striatal axis, it is unclear how distinct fronto-striatal sub-circuits support value-based choice. Here we found segregated prefrontal populations defined by anterior/posterior dorsomedial striatal target. During a feedback-based 2-alternative choice task, single-photon imaging revealed circuit-specific representations of task-relevant information with prelimbic neurons targeting anterior DMS (PL::A-DMS) robustly modulated during choices and negative outcomes, while prelimbic neurons targeting posterior DMS (PL::P-DMS) encoded internal representations of value and positive outcomes contingent on prior choice. Consistent with this distributed coding, optogenetic inhibition of PL::A-DMS circuits strongly impacted choice monitoring and responses to negative outcomes while inhibition of PL::P-DMS impaired task engagement and strategies following positive outcomes. Together our data uncover PL populations engaged in distributed processing for value-based choice.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Temporally and Functionally Distinct Contributions to Value Based Choice Along the Anterior-Posterior Dorsomedial Striatal Axis.bioRxiv [Preprint]. 2025 Mar 15:2025.03.14.643367. doi: 10.1101/2025.03.14.643367. bioRxiv. 2025. PMID: 40161810 Free PMC article. Preprint.
-
Differential encoding of action selection by orbitofrontal and striatal population dynamics.J Neurophysiol. 2020 Aug 1;124(2):634-644. doi: 10.1152/jn.00316.2020. Epub 2020 Jul 29. J Neurophysiol. 2020. PMID: 32727312 Free PMC article.
-
Distinct Medial Orbitofrontal-Striatal Circuits Support Dissociable Component Processes of Risk/Reward Decision-Making.J Neurosci. 2022 Mar 30;42(13):2743-2755. doi: 10.1523/JNEUROSCI.2097-21.2022. Epub 2022 Feb 8. J Neurosci. 2022. PMID: 35135853 Free PMC article.
-
Parallel associative processing in the dorsal striatum: segregation of stimulus-response and cognitive control subregions.Neurobiol Learn Mem. 2011 Sep;96(2):95-120. doi: 10.1016/j.nlm.2011.06.002. Epub 2011 Jun 16. Neurobiol Learn Mem. 2011. PMID: 21704718 Review.
-
Involvement of basal ganglia and orbitofrontal cortex in goal-directed behavior.Prog Brain Res. 2000;126:193-215. doi: 10.1016/S0079-6123(00)26015-9. Prog Brain Res. 2000. PMID: 11105648 Review.
Cited by
-
Stochastic characterization of navigation strategies in an automated variant of the Barnes maze.Elife. 2024 Jun 20;12:RP88648. doi: 10.7554/eLife.88648. Elife. 2024. PMID: 38899521 Free PMC article.
-
An Open-Source Joystick Platform for Investigating Forelimb Motor Control, Auditory-Motor Integration, and Value-Based Decision-Making in Head-Fixed Mice.eNeuro. 2025 Apr 28;12(4):ENEURO.0038-25.2025. doi: 10.1523/ENEURO.0038-25.2025. Print 2025 Apr. eNeuro. 2025. PMID: 40295100 Free PMC article.
-
Cortico-striatal action control inherent of opponent cognitive-motivational styles.bioRxiv [Preprint]. 2024 Dec 5:2024.03.12.584623. doi: 10.1101/2024.03.12.584623. bioRxiv. 2024. Update in: Elife. 2025 Feb 19;13:RP100988. doi: 10.7554/eLife.100988. PMID: 38559086 Free PMC article. Updated. Preprint.
-
A feature-specific prediction error model explains dopaminergic heterogeneity.Nat Neurosci. 2024 Aug;27(8):1574-1586. doi: 10.1038/s41593-024-01689-1. Epub 2024 Jul 3. Nat Neurosci. 2024. PMID: 38961229
-
Striatal arbitration between choice strategies guides few-shot adaptation.Nat Commun. 2025 Feb 20;16(1):1811. doi: 10.1038/s41467-025-57049-5. Nat Commun. 2025. PMID: 39979316 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

