Melatonin inhibits bladder tumorigenesis by suppressing PPARγ/ENO1-mediated glycolysis
- PMID: 37024456
- PMCID: PMC10079981
- DOI: 10.1038/s41419-023-05770-8
Melatonin inhibits bladder tumorigenesis by suppressing PPARγ/ENO1-mediated glycolysis
Abstract
Melatonin is a well-known natural hormone, which shows a potential anticancer effect in many human cancers. Bladder cancer (BLCA) is one of the most malignant human cancers in the world. Chemoresistance is an increasingly prominent phenomenon that presents an obstacle to the clinical treatment of BLCA. There is an urgent need to investigate novel drugs to improve the current clinical status. In our study, we comprehensively explored the inhibitory effect of melatonin on BLCA and found that it could suppress glycolysis process. Moreover, we discovered that ENO1, a glycolytic enzyme involved in the ninth step of glycolysis, was the downstream effector of melatonin and could be a predictive biomarker of BLCA. We also proved that enhanced glycolysis simulated by adding exogenous pyruvate could induce gemcitabine resistance, and melatonin treatment or silencing of ENO1 could intensify the cytotoxic effect of gemcitabine on BLCA cells. Excessive accumulation of reactive oxygen species (ROS) mediated the inhibitory effect of melatonin on BLCA cells. Additionally, we uncovered that PPARγ was a novel upstream regulator of ENO1, which mediated the downregulation of ENO1 caused by melatonin. Our study offers a fresh perspective on the anticancer effect of melatonin and encourages further studies on clinical chemoresistance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


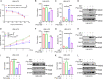
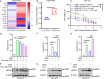


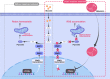
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

