Factor Xa cleaves SARS-CoV-2 spike protein to block viral entry and infection
- PMID: 37024459
- PMCID: PMC10079155
- DOI: 10.1038/s41467-023-37336-9
Factor Xa cleaves SARS-CoV-2 spike protein to block viral entry and infection
Abstract
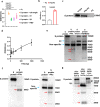
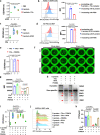
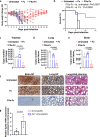
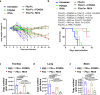
Serine proteases (SP), including furin, trypsin, and TMPRSS2 cleave the SARS-CoV-2 spike (S) protein, enabling the virus to enter cells. Here, we show that factor (F) Xa, an SP involved in blood coagulation, is upregulated in COVID-19 patients. In contrast to other SPs, FXa exerts antiviral activity. Mechanistically, FXa cleaves S protein, preventing its binding to ACE2, and thus blocking viral entry and infection. However, FXa is less effective against variants carrying the D614G mutation common in all pandemic variants. The anticoagulant rivaroxaban, a direct FXa inhibitor, inhibits FXa-mediated S protein cleavage and facilitates viral entry, whereas the indirect FXa inhibitor fondaparinux does not. In the lethal SARS-CoV-2 K18-hACE2 model, FXa prolongs survival yet its combination with rivaroxaban but not fondaparinux abrogates that protection. These results identify both a previously unknown function for FXa and an associated antiviral host defense mechanism against SARS-CoV-2 and suggest caution in considering direct FXa inhibitors for preventing or treating thrombotic complications in COVID-19 patients.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

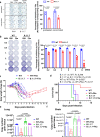
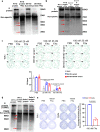
References
-
- Worldometer. Worldometer’s COVID-19 data. <https://www.worldometers.info/coronavirus/country/us/> (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

