Replication-associated formation and repair of human topoisomerase IIIα cleavage complexes
- PMID: 37024461
- PMCID: PMC10079683
- DOI: 10.1038/s41467-023-37498-6
Replication-associated formation and repair of human topoisomerase IIIα cleavage complexes
Abstract
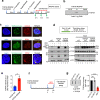
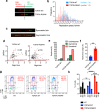
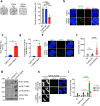
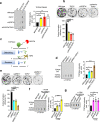
Topoisomerase IIIα (TOP3A) belongs to the conserved Type IA family of DNA topoisomerases. Here we report that human TOP3A is associated with DNA replication forks and that a "self-trapping" TOP3A mutant (TOP3A-R364W) generates cellular TOP3A DNA cleavage complexes (TOP3Accs). We show that trapped TOP3Accs that interfere with replication, induce DNA damage and genome instability. To elucidate how TOP3Accs are repaired, we explored the role of Spartan (SPRTN), the metalloprotease associated with DNA replication, which digests proteins forming DNA-protein crosslinks (DPCs). We find that SPRTN-deficient cells show elevated TOP3Accs, whereas overexpression of SPRTN lowers cellular TOP3Accs. SPRTN is deubiquitinated and epistatic with TDP2 in response to TOP3Accs. In addition, we found that MRE11 can excise TOP3Accs, and that cell cycle determines the preference for the SPRTN-TDP2 vs. the ATM-MRE11 pathways, in S vs. G2, respectively. Our study highlights the prevalence of TOP3Accs repair mechanisms to ensure normal DNA replication.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
TOP3A coupling with replication forks and repair of TOP3A cleavage complexes.Cell Cycle. 2024 Jan;23(2):115-130. doi: 10.1080/15384101.2024.2314440. Epub 2024 Feb 11. Cell Cycle. 2024. PMID: 38341866 Free PMC article. Review.
-
PARP1-driven repair of topoisomerase IIIα DNA-protein crosslinks by FEN1.Cell Rep. 2024 Aug 27;43(8):114522. doi: 10.1016/j.celrep.2024.114522. Epub 2024 Jul 18. Cell Rep. 2024. PMID: 39028621 Free PMC article.
-
SPRTN and TDP1/TDP2 Independently Suppress 5-Aza-2'-deoxycytidine-Induced Genomic Instability in Human TK6 Cell Line.Chem Res Toxicol. 2022 Nov 21;35(11):2059-2067. doi: 10.1021/acs.chemrestox.2c00213. Epub 2022 Oct 25. Chem Res Toxicol. 2022. PMID: 36282523
-
Tyrosyl-DNA phosphodiesterase 1 (TDP1) and SPRTN protease repair histone 3 and topoisomerase 1 DNA-protein crosslinks in vivo.Open Biol. 2023 Oct;13(10):230113. doi: 10.1098/rsob.230113. Epub 2023 Oct 4. Open Biol. 2023. PMID: 37788708 Free PMC article.
-
From the TOP: Formation, recognition and resolution of topoisomerase DNA protein crosslinks.DNA Repair (Amst). 2024 Oct;142:103751. doi: 10.1016/j.dnarep.2024.103751. Epub 2024 Aug 16. DNA Repair (Amst). 2024. PMID: 39180935 Review.
Cited by
-
Application of a bivalent "click" approach to target tyrosyl-DNA phosphodiesterase 1 (TDP1).RSC Med Chem. 2025 Feb 21;16(5):1969-1985. doi: 10.1039/d4md00824c. eCollection 2025 May 22. RSC Med Chem. 2025. PMID: 39990162 Free PMC article.
-
Poly(ADP-ribosyl)ation of TIMELESS limits DNA replication stress and promotes stalled fork protection.Cell Rep. 2024 Mar 26;43(3):113845. doi: 10.1016/j.celrep.2024.113845. Epub 2024 Feb 22. Cell Rep. 2024. PMID: 38393943 Free PMC article.
-
Flap endonuclease 1 repairs DNA-protein cross-links via ADP-ribosylation-dependent mechanisms.Sci Adv. 2025 Jan 10;11(2):eads2919. doi: 10.1126/sciadv.ads2919. Epub 2025 Jan 10. Sci Adv. 2025. PMID: 39792662 Free PMC article.
-
Deciphering the human TopIIIα activity modulated by Rmi1 using magnetic tweezers.Nucleic Acids Res. 2025 Apr 22;53(8):gkaf308. doi: 10.1093/nar/gkaf308. Nucleic Acids Res. 2025. PMID: 40266687 Free PMC article.
-
Temporospatial control of topoisomerases by essential cellular processes.Curr Opin Microbiol. 2024 Dec;82:102559. doi: 10.1016/j.mib.2024.102559. Epub 2024 Nov 8. Curr Opin Microbiol. 2024. PMID: 39520813 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

