Tryptase β regulation of joint lubrication and inflammation via proteoglycan-4 in osteoarthritis
- PMID: 37024468
- PMCID: PMC10079686
- DOI: 10.1038/s41467-023-37598-3
Tryptase β regulation of joint lubrication and inflammation via proteoglycan-4 in osteoarthritis
Abstract
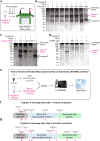
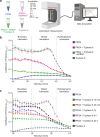
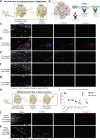
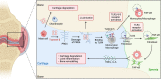
PRG4 is an extracellular matrix protein that maintains homeostasis through its boundary lubricating and anti-inflammatory properties. Altered expression and function of PRG4 have been associated with joint inflammatory diseases, including osteoarthritis. Here we show that mast cell tryptase β cleaves PRG4 in a dose- and time-dependent manner, which was confirmed by silver stain gel electrophoresis and mass spectrometry. Tryptase-treated PRG4 results in a reduction of lubrication. Compared to full-length, cleaved PRG4 further activates NF-κB expression in cells overexpressing TLR2, -4, and -5. In the destabilization of the medial meniscus model of osteoarthritis in rat, tryptase β and PRG4 colocalize at the site of injury in knee cartilage and is associated with disease severity. When human primary synovial fibroblasts from male osteoarthritis patients or male healthy subjects treated with tryptase β and/or PRG4 are subjected to a quantitative shotgun proteomics and proteome changes are characterized, it further supports the role of NF-κB activation. Here we show that tryptase β as a modulator of joint lubrication in osteoarthritis via the cleavage of PRG4.
© 2023. Crown.
Conflict of interest statement
T.A.S. and G.D.J. have authored patents on rhPRG4 and hold equity in Lubris LLC, MA, USA. T.A.S. is also a paid consultant for Lubris LLC, MA, USA. R.K. also has authored patents on rhPRG4. All other authors have nothing to disclose.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

