A roadmap to establish a comprehensive platform for sustainable manufacturing of natural products in yeast
- PMID: 37024483
- PMCID: PMC10079933
- DOI: 10.1038/s41467-023-37627-1
A roadmap to establish a comprehensive platform for sustainable manufacturing of natural products in yeast
Abstract
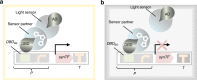
Secondary natural products (NPs) are a rich source for drug discovery. However, the low abundance of NPs makes their extraction from nature inefficient, while chemical synthesis is challenging and unsustainable. Saccharomyces cerevisiae and Pichia pastoris are excellent manufacturing systems for the production of NPs. This Perspective discusses a comprehensive platform for sustainable production of NPs in the two yeasts through system-associated optimization at four levels: genetics, temporal controllers, productivity screening, and scalability. Additionally, it is pointed out critical metabolic building blocks in NP bioengineering can be identified through connecting multilevel data of the optimized system using deep learning.
© 2023. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures



Similar articles
-
High-Throughput Indirect Monitoring of TORC1 Activation Using the pTOMAN-G Plasmid in Yeast.Bio Protoc. 2025 Jun 20;15(12):e5356. doi: 10.21769/BioProtoc.5356. eCollection 2025 Jun 20. Bio Protoc. 2025. PMID: 40620813 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
Cited by
-
De novo-assembled long-read genomes of Saccharomyces cerevisiae strains in commerce.Microbiol Resour Announc. 2025 Jan 16;14(1):e0070524. doi: 10.1128/mra.00705-24. Epub 2024 Dec 12. Microbiol Resour Announc. 2025. PMID: 39665563 Free PMC article.
-
Introducing carbon assimilation in yeasts using photosynthetic directed endosymbiosis.Nat Commun. 2024 Jul 16;15(1):5947. doi: 10.1038/s41467-024-49585-3. Nat Commun. 2024. PMID: 39013857 Free PMC article.
-
Creation of a eukaryotic multiplexed site-specific inversion system and its application for metabolic engineering.Nat Commun. 2025 Feb 24;16(1):1918. doi: 10.1038/s41467-025-57206-w. Nat Commun. 2025. PMID: 39994248 Free PMC article.
-
General mechanisms of weak acid-tolerance and current strategies for the development of tolerant yeasts.World J Microbiol Biotechnol. 2023 Dec 22;40(2):49. doi: 10.1007/s11274-023-03875-y. World J Microbiol Biotechnol. 2023. PMID: 38133718 Review.
-
Combinatorial optimization of gene expression through recombinase-mediated promoter and terminator shuffling in yeast.Nat Commun. 2024 Feb 7;15(1):1112. doi: 10.1038/s41467-024-44997-7. Nat Commun. 2024. PMID: 38326309 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

