Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species
- PMID: 37024581
- PMCID: PMC10181942
- DOI: 10.1038/s41588-023-01340-y
Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species
Abstract
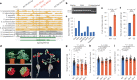
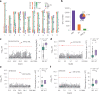
Effective utilization of wild relatives is key to overcoming challenges in genetic improvement of cultivated tomato, which has a narrow genetic basis; however, current efforts to decipher high-quality genomes for tomato wild species are insufficient. Here, we report chromosome-scale tomato genomes from nine wild species and two cultivated accessions, representative of Solanum section Lycopersicon, the tomato clade. Together with two previously released genomes, we elucidate the phylogeny of Lycopersicon and construct a section-wide gene repertoire. We reveal the landscape of structural variants and provide entry to the genomic diversity among tomato wild relatives, enabling the discovery of a wild tomato gene with the potential to increase yields of modern cultivated tomatoes. Construction of a graph-based genome enables structural-variant-based genome-wide association studies, identifying numerous signals associated with tomato flavor-related traits and fruit metabolites. The tomato super-pangenome resources will expedite biological studies and breeding of this globally important crop.
© 2023. The Author(s).
Conflict of interest statement
N.L., Q.H., B.W., J.W., Q.Y., T.Y. and P.A. have filed patent applications on technology related to the processes described in this article (Chinese patent application number CN202111231381.0; ‘Application of the
Figures


References
-
- Tieman D, et al. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. - PubMed
-
- Peralta IE, Spooner DM, Knapp S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae) Syst. Bot. Monogr. 2008;84:1–186.
-
- Rick, C. M. Perspectives from plant genetics: the Tomato Genetics Stock Center. In Genetic resources at risk: scientific issues, technologies, and funding policies. Proceedings of a symposium, American Association for the Advancement of Science annual meeting, San Francisco, California, USA, 16 January 1989 (Eds McGuire, P. E. & Qualset, C. O.) 11–19 (Genetic Resources Conservation Program, University of California, 1990).
-
- Mutschler MA, et al. QTL analysis of pest resistance in the wild tomato Lycopersicon pennellii: QTLs controlling acylsugar level and composition. Theor. Appl. Genet. 1996;92:709–718. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

