Genomic and transcriptomic analysis of checkpoint blockade response in advanced non-small cell lung cancer
- PMID: 37024582
- PMCID: PMC10181943
- DOI: 10.1038/s41588-023-01355-5
Genomic and transcriptomic analysis of checkpoint blockade response in advanced non-small cell lung cancer
Abstract
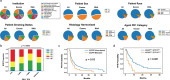
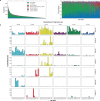
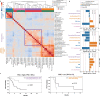
Anti-PD-1/PD-L1 agents have transformed the treatment landscape of advanced non-small cell lung cancer (NSCLC). To expand our understanding of the molecular features underlying response to checkpoint inhibitors in NSCLC, we describe here the first joint analysis of the Stand Up To Cancer-Mark Foundation cohort, a resource of whole exome and/or RNA sequencing from 393 patients with NSCLC treated with anti-PD-(L)1 therapy, along with matched clinical response annotation. We identify a number of associations between molecular features and outcome, including (1) favorable (for example, ATM altered) and unfavorable (for example, TERT amplified) genomic subgroups, (2) a prominent association between expression of inducible components of the immunoproteasome and response and (3) a dedifferentiated tumor-intrinsic subtype with enhanced response to checkpoint blockade. Taken together, results from this cohort demonstrate the complexity of biological determinants underlying immunotherapy outcomes and reinforce the discovery potential of integrative analysis within large, well-curated, cancer-specific cohorts.
© 2023. The Author(s).
Conflict of interest statement
A.R. is a founder, equity owner, and consultant at Halo Solutions and has served as a consultant at Tyra Biosciences. J.F.G. has served as a compensated consultant or received honoraria from Bristol Myers Squibb, Genentech/Roche, Ariad/Takeda, Loxo/Lilly, Blueprint, Oncorus, Regeneron, Gilead, Moderna, Mirati, AstraZeneca, Pfizer, Novartis, iTeos, Nuvalent, Karyopharm, Beigene, Silverback Therapeutics, Merck and GlydeBio; research support from Novartis, Genentech/Roche and Ariad/Takeda; institutional research support from Bristol Myers Squibb, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis and Alexo; and has an immediate family member who is an employee with equity at Ironwood Pharmaceuticals. S.S.F is an inventor on provisional patent application No. 62/866,261 related to methods for predicting outcomes of checkpoint inhibition in melanoma and his salary was partially supported by research funding from IBM. I.L. owns equity and consults for ennov1, LLC, and additionally consults for PACT Pharma. J.K. is a current employee and equity owner of GlaxoSmithKline. N.I.V. is a consultant for Sanofi/Regeneron, Oncocyte, and Lilly. P.M.F. has served as a consultant for Amgen, AstraZeneca, BMS, Daichii, F-Star, G1, Genentech, Iteos, Janssen, Novartis, Sanofi, and Surface and has received research support from AstraZeneca, Biontech, BMS, and Novartis. V.A. has received research support to Johns Hopkins from Bristol Myers Squibb and AstraZeneca. J.W.R. has served as a consultant for Boehringer Ingelheim, Novartis, Blueprint, Daiichi Sankyo, EMD Serano, Jazz Pharmaceuticals, Bristol Myers Squibb, Janssen Oncology, Beigene, Turning Point Therapeutics, Genentech and receives research funding from AstraZeneca, Spectrum, Merck, Boehringer Ingelheim, Novartis, Revolution Medicines, GlaxoSmithKline. D.L.G. is an equity owner in Exact Sciences and Nektar; consults for Sanofi, GlaxoSmithKline, Alethia Biotherapeutics, Janssen Research & Development, Eli Lilly, Menarini Ricerche, and 4D Pharma; and receives research support from Janssen Research & Development, Takeda, AstraZeneca, Astellas, Ribon Therapeutics, and NGM Biopharmaceuticals. N.A.P. is a consultant for Astrazeneca, Merck, Pfizer, Eli Lilly/LOXO, Genentech, BMS, Amgen, Mirati, Inivata, G1 Therapeutics, Viosera, Xencor, Janssen, and Boehringer Ingelheim and receives research funding from LOXO, BMS, Merck, Heat Bio, WindMIL, Genentech, Astrazeneca, Spectrum, Mirati, Altor, Jounce, and Sanofi. V.V. is a consultant for BMS, Merck, AstraZeneca, Foundation Medicine, Novartis, Iteos Therapeutics, EMD Serono and receives research funding from AstraZeneca. S.R.D. provides independent image analysis for hospital-contracted clinical research trials programs for Merck, Pfizer, Bristol Myers Squibb, Novartis, Roche, Polaris, Cascadian, Abbvie, Gradalis, Bayer, Zai laboratories, Biengen, Resonance, and Analise and receives research support from Lunit Inc, GE, Vuno and Qure AI. M.M. is a consultant for AstraZeneca, H3 Biomedicine, BMS, Sanofi, Janssen Oncology; receives research funding from Novartis; and owns intellectual property in Elsevier. A.C. is a founder, equity holder, and consultant of Darwin Health Inc. (Columbia University is also an equity holder); holds intellectual property in US patent number 10,790,040 has been awarded related to this work, assigned to Columbia University with A.C. as an inventor, and US patent application number 20210327537 has been filed, also for assignment to Columbia University with A.C. as an inventor. J.V.H. is a consultant for AstraZeneca, BioCurity Pharmaceuticals, Boehringer Ingelheim Pharma, Bristol Myers Squibb, Chugai Biopharmaceuticals, Eli Lilly & Co, EMD Serono, Inc., Genentech, Janssen, Mirati Therapeutics, OncoCyte, Reflexion, Regeneron Pharmaceuticals, Sandoz Pharmaceuticals, Sanofi US Services, Takeda, uniQure, DAVA Oncology, BrightPath Biotherapeutics, Pneuma Respiratory, Eisai, Kairos Venture Investments, GlaxoSmithKline, Gritstone Oncology, Targeted Oncology, Intellisphere, LLC, Millennium Pharmaceuticals, Inc., Catalyst Pharmaceuticals, Guardant Health, Inc., Hengrui Therapeutics, Inc., and Leads Biolabs; receives research funding from AstraZeneca, GlaxoSmithKline, Spectrum; and has intellectual property in Spectrum. R.S.H. has equity in Immunocore, and Bolt, Checkpoint Therapeutics; consults for Immunocore, Junshi Pharmaceuticals, Abbvie, ARMO, AstraZeneca, Bayer, Bolt, Bristol Myers Squibb, Candel Therapeutics, Cybrexa Therapeutics, DynamiCure Biotechnology, eFFECTOR Therapeutics, Eli Lilly, EMD Serono, Foundation Medicine, Genentech/Roche, Genmab, Gliead, Halozyme, Heat Biologics, HiberCell, I-Mab Biopharma, Immune-Onc Therapeutics, Immunocore, Infinity Pharmaceuticals, Johnson and Johnson, Loxo Oncology, Merck, Mirati Therapeutics, Nektar, Neon Therapeutics, NextCure, Novartis, Ocean Biomedical, Oncocyte Corp, Oncternal Therapeutics, Pfizer, Refactor Health, Ribbon Therapeutics, Sanofi, Seattle Genetics, Shire PLC, Spectrum, STCube, Symphogen, Takeda, Tesaro, Tocagen, Ventana Medical Systems, WindMIL Therapeutics, and Xencor; and receives research support from AstraZeneca, Eli Lilly, Genentech/Roche, and Merck. J.R.B. is a consultant for Amgen, Johnson & Johnson, Merck, Bristol Myers Squibb, Sanofi, GlaxoSmithKline, Janssen, Bluprint, AstraZeneca, Regeneron, and Eli Lilly and receives research funding from Bristol Myers Squibb. K.A.S. is a consultant for Shattuck Labs, Pierre-Fabre, EMD Serono, Clinica Alemana de Santiago, Genmab, Takeda, Merck Sharpe & Dohme, Bristol Myers Squibb, AstraZeneca, Agenus and Torque Therapeutics and receives research funding from Navigate Biopharma, Tesaro/GSK, Moderna Inc., Takeda, Surface Oncology, Pierre-Fabre Research Institute, Merck Sharpe & Dohme, Bristol Myers Squibb, AstraZeneca, Ribon Therapeutics, Akoya Biosciences, Boehringer Ingelheim and Eli Lilly. V.E.V. is a founder of Delfi Diagnostics, serves as on the Board of Directors and as a consultant for this organization, and owns Delfi Diagnostics stock, which is subject to certain restrictions under university policy. Additionally, Johns Hopkins University owns equity in Delfi Diagnostics. V.E.V. divested his equity in Personal Genome Diagnostics (PGDx) to LabCorp in February 2022. V.E.V. is an inventor on patent applications submitted by Johns Hopkins University related to cancer genomic analyses and cell-free DNA for cancer detection that have been licensed to one or more entities, including Delfi Diagnostics, LabCorp, Qiagen, Sysmex, Agios, Genzyme, Esoterix, Ventana and ManaT Bio. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. V.E.V. is an advisor to Viron Therapeutics and Epitope. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. N.A.R. is an equity owner in Synthekine and Gritstone; holds positions as CMO of Synthekine and member of Board of Directors and Scientific Advisory Board of Gristone; and holds intellectual property related to Determinants of cancer response to immunotherapy (PCT/US2015/062208) licensed to Personal Genome Diagnostics. P.A.J. is an equity owner in Gatekeeper Pharmaceuticals; consults for AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Chugai Pharmaceuticals, Eli Lilly Pharmaceuticals, Araxes Pharmaceuticals, SFJ Pharmaceuticals, Voronoi, Daiichi Sankyo, Biocartis, Novartis, Sanofi, Takeda Oncology, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Esai, Allorion Therapeutics, Accutar Biotech, and Abbvie; receives research support from AstraZeneca, Daiichi Sankyo, PUMA, Eli Lilly, Boehringer Ingelheim, Revolution Medicines, and Takeda Oncology and is a co-inventor and receives postmarketing royalties on a DFCI owned patent on EGFR mutations licensed to LabCorp. M.M.A. is a consultant for Genentech, Bristol Myers Squibb, Merck, AstraZeneca, AbbVie, Neon, Achilles, Maverick, Blueprint Medicine, Hengrui, Syndax, Ariad, Nektar, Gritstone, ArcherDX, Mirati, NextCure, Novartis, EMD Serono, Panvaxal/NovaRx, and Foundation Medicine; and is supported by research grants from Genentech, Lilly, Bristol Myers Squibb, and AstraZeneca. B.D.G. is an equity owner in Rome Therapeutics; consults for Darwin Health, Merck, PMV Pharma and Rome Therapeutics; has received research funding for Bristol Myers Squibb and Merck; and is an owner of intellectual property related to: Rna containing compositions and methods of their use (WO2016131048A1), Neoantigens and uses thereof for treating cancer (US20200232040A1), and Compositions and methods for inhibiting cancers and viruses (WO2020023776A2). M.L. is an owner of intellectual property related to: Neoantigens and uses thereof for treating cancer (WO2018136664A1). A.T.S. is an equity owner and current employee of Novartis. J.W. has equity in Tizona Pharmaceuticals, Imvaq, Beigene, Linneaus, Apricity, Arsenal IO, Georgiamune, Trieza, Maverick, Ascentage; consults for Amgen, Apricity, Ascentage Pharma, Arsenal IO, Astellas, AstraZeneca, Bayer, Bicara Therapeutics, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, Dragonfly, Eli Lilly, F Star, Georgiamune, Idera, Imvaq, Maverick Therapeutics, Merck, Psioxus, Recepta, Tizona, Trieza, Truvax, Trishula, Sellas, Surface Oncology, Syndax, Syntalogic, Werewolf Therapeutics; receives research support from Bristol Myers Squibb and Sephora; and owns intellectual property related to: Xenogeneic DNA Vaccines, Alphavirus replicon particles expressing TRP2, Myeloid-derived suppressor cell (MDSC) assay, Newcastle Disease viruses for Cancer Therapy, Vaccinia virus mutants useful for cancer immunotherapy, Anti-PD-1 Antibody, Anti-CTLA4 antibodies, Anti-GITR antibodies and methods of use thereof, Identifying And Treating Subjects At Risk For Checkpoint Blockade Therapy Associated Colitis, Immunosuppressive follicular helper-like T cells modulated by immune checkpoint blockade, CD40 binding molecules and uses thereof, Phosphatidylserine Targeting Agents and uses thereof for adoptive T-cell therapies, Anti-CD40 agonist mAb fused to Monophosphoryl Lipid A (MPL) for cancer therapy, CAR+T cells targeting differentiation antigens as means to treat cancer. N.H. is an equity owner in BioNtech, Related Sciences/Danger Bio, and consults for Related Sciences/Danger Bio. G.G. is an equity holder in Scorpion Therapeutics; consults for Scorpion Therapeutics; receives research funding from IBM and Pharmacyclics; and is an inventor on patent applications related to MSMuTect, MSMutSig, MSIDetect, POLYSOLVER, SignatureAnalyzer-GPU and TensorQTL. M.D.H. has equity in Factorial, Immunai, Shattuck Labs, Arcus, and Avail Bio, and began as an employee and equity holder at AstraZeneca subsequent to the completion of this work; has consulted for Achilles, Adagene, Adicet, Arcus, AstraZeneca, Blueprint, BMS, DaVolterra, Eli Lilly, Genentech/Roche, Genzyme/Sanofi, Janssen, Immunai, Instil Bio, Mana Therapeutics, Merck, Mirati, Natera, Pact Pharma, Shattuck Labs, and Regeneron; has received research funding from Bristol Myers Squibb; and has intellectual property regarding a patent filed by Memorial Sloan Kettering related to the use of TMB to predict response to immunotherapy (PCT/US2015/062208), which is pending and licensed by PGDx. The remaining authors declare no competing interests.
Figures










References
-
- Herbst RS, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540–1550. - PubMed
-
- Reck M, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. - PubMed
-
- Paz-Ares L, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N. Engl. J. Med. 2018;379:2040–2051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA093373/CA/NCI NIH HHS/United States
- U01 CA272610/CA/NCI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- T32 GM007367/GM/NIGMS NIH HHS/United States
- F30 CA257765/CA/NCI NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- S10 OD032433/OD/NIH HHS/United States
- T32 CA009172/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- S10 OD012351/OD/NIH HHS/United States
- K12 CA138464/CA/NCI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- U01 CA217858/CA/NCI NIH HHS/United States
- R01 CA240317/CA/NCI NIH HHS/United States
- R01 CA208756/CA/NCI NIH HHS/United States
- S10 OD021764/OD/NIH HHS/United States
- R35 CA197745/CA/NCI NIH HHS/United States
- K08 CA248723/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

