Chemical and genomic characterization of a potential probiotic treatment for stony coral tissue loss disease
- PMID: 37024599
- PMCID: PMC10079959
- DOI: 10.1038/s42003-023-04590-y
Chemical and genomic characterization of a potential probiotic treatment for stony coral tissue loss disease
Abstract
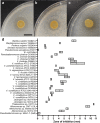
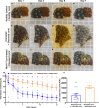
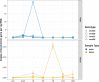
Considered one of the most devastating coral disease outbreaks in history, stony coral tissue loss disease (SCTLD) is currently spreading throughout Florida's coral reefs and the greater Caribbean. SCTLD affects at least two dozen different coral species and has been implicated in extensive losses of coral cover. Here we show Pseudoalteromonas sp. strain McH1-7 has broad-spectrum antibacterial activity against SCTLD-associated bacterial isolates. Chemical analyses indicated McH1-7 produces at least two potential antibacterials, korormicin and tetrabromopyrrole, while genomic analysis identified the genes potentially encoding an L-amino acid oxidase and multiple antibacterial metalloproteases (pseudoalterins). During laboratory trials, McH1-7 arrested or slowed disease progression on 68.2% of diseased Montastraea cavernosa fragments treated (n = 22), and it prevented disease transmission by 100% (n = 12). McH1-7 is the most chemically characterized coral probiotic that is an effective prophylactic and direct treatment for the destructive SCTLD as well as a potential alternative to antibiotic use.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Knowlton, N. et al. Rebuilding coral reefs: a decadal grand challenge. 1–55 10.53642/NRKY9386 (2021).
-
- Walton, C. J., Hayes, N. K. & Gilliam, D. S. Impacts of a regional, multi-year, multi-species coral disease outbreak in Southeast Florida. Front. Mar. Sci.5, 323 (2018).
-
- Spadafore R, Fura R, Precht WF, Vollmer SV. Multi-variate analyses of coral mortality from the 2014–2015 stony coral tissue loss disease outbreak off Miami-Dade county, Florida. Front. Mar. Sci. 2021;8:723998. doi: 10.3389/fmars.2021.723998. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

