Viruses interact with hosts that span distantly related microbial domains in dense hydrothermal mats
- PMID: 37024618
- PMCID: PMC10159854
- DOI: 10.1038/s41564-023-01347-5
Viruses interact with hosts that span distantly related microbial domains in dense hydrothermal mats
Abstract
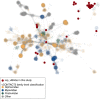
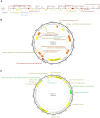
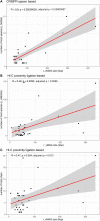
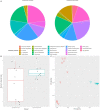
Many microbes in nature reside in dense, metabolically interdependent communities. We investigated the nature and extent of microbe-virus interactions in relation to microbial density and syntrophy by examining microbe-virus interactions in a biomass dense, deep-sea hydrothermal mat. Using metagenomic sequencing, we find numerous instances where phylogenetically distant (up to domain level) microbes encode CRISPR-based immunity against the same viruses in the mat. Evidence of viral interactions with hosts cross-cutting microbial domains is particularly striking between known syntrophic partners, for example those engaged in anaerobic methanotrophy. These patterns are corroborated by proximity-ligation-based (Hi-C) inference. Surveys of public datasets reveal additional viruses interacting with hosts across domains in diverse ecosystems known to harbour syntrophic biofilms. We propose that the entry of viral particles and/or DNA to non-primary host cells may be a common phenomenon in densely populated ecosystems, with eco-evolutionary implications for syntrophic microbes and CRISPR-mediated inter-population augmentation of resilience against viruses.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
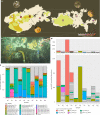

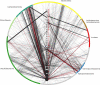
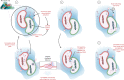
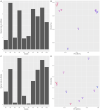




Similar articles
-
Deep-Sea Hydrothermal Vent Viruses Compensate for Microbial Metabolism in Virus-Host Interactions.mBio. 2017 Jul 11;8(4):e00893-17. doi: 10.1128/mBio.00893-17. mBio. 2017. PMID: 28698277 Free PMC article.
-
Unveiling Ecological and Genetic Novelty within Lytic and Lysogenic Viral Communities of Hot Spring Phototrophic Microbial Mats.Microbiol Spectr. 2021 Dec 22;9(3):e0069421. doi: 10.1128/Spectrum.00694-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787442 Free PMC article.
-
Virus diversity and interactions with hosts in deep-sea hydrothermal vents.Microbiome. 2022 Dec 24;10(1):235. doi: 10.1186/s40168-022-01441-6. Microbiome. 2022. PMID: 36566239 Free PMC article.
-
Ecological and evolutionary interactions in syntrophic methanogenic consortia.Microbes Environ. 2010;25(3):145-51. doi: 10.1264/jsme2.me10122. Microbes Environ. 2010. PMID: 21576866 Review.
-
Between a Rock and a Soft Place: The Role of Viruses in Lithification of Modern Microbial Mats.Trends Microbiol. 2021 Mar;29(3):204-213. doi: 10.1016/j.tim.2020.06.004. Epub 2020 Jul 9. Trends Microbiol. 2021. PMID: 32654857 Review.
Cited by
-
Virus-host interactions predictor (VHIP): Machine learning approach to resolve microbial virus-host interaction networks.PLoS Comput Biol. 2024 Sep 18;20(9):e1011649. doi: 10.1371/journal.pcbi.1011649. eCollection 2024 Sep. PLoS Comput Biol. 2024. PMID: 39292721 Free PMC article.
-
Hot springs viruses at Yellowstone National Park have ancient origins and are adapted to thermophilic hosts.Commun Biol. 2024 Apr 9;7(1):312. doi: 10.1038/s42003-024-05931-1. Commun Biol. 2024. PMID: 38594478 Free PMC article.
-
Host-specific viral predation network on coral reefs.ISME J. 2024 Jan 8;18(1):wrae240. doi: 10.1093/ismejo/wrae240. ISME J. 2024. PMID: 39657233 Free PMC article.
-
Synthetic community Hi-C benchmarking provides a baseline for virus-host inferences.bioRxiv [Preprint]. 2025 Apr 23:2025.02.12.637985. doi: 10.1101/2025.02.12.637985. bioRxiv. 2025. PMID: 39990352 Free PMC article. Preprint.
-
Decoupling of strain- and intrastrain-level interactions of microbiomes in a sponge holobiont.Nat Commun. 2024 Sep 18;15(1):8205. doi: 10.1038/s41467-024-52464-6. Nat Commun. 2024. PMID: 39294150 Free PMC article.

