The selection of a hydrophobic 7-phenylbutyl-7-deazaadenine-modified DNA aptamer with high binding affinity for the Heat Shock Protein 70
- PMID: 37024672
- PMCID: PMC10079658
- DOI: 10.1038/s42004-023-00862-0
The selection of a hydrophobic 7-phenylbutyl-7-deazaadenine-modified DNA aptamer with high binding affinity for the Heat Shock Protein 70
Abstract
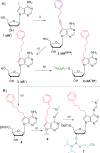
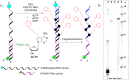
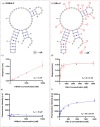
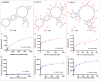
Nucleic acids aptamers often fail to efficiently target some proteins because of the hydrophilic character of the natural nucleotides. Here we present hydrophobic 7-phenylbutyl-7-deaadenine-modified DNA aptamers against the Heat Shock Protein 70 that were selected via PEX and magnetic bead-based SELEX. After 9 rounds of selection, the pool was sequenced and a number of candidates were identified. Following initial screening, two modified aptamers were chemically synthesised in-house and their binding affinity analysed by two methods, bio-layer interferometry and fluorescent-plate-based binding assay. The binding affinities of the modified aptamers were compared with that of their natural counterparts. The resulting modified aptamers bound with higher affinity (low nanomolar range) to the Hsp70 than their natural sequence (>5 µM) and hence have potential for applications and further development towards Hsp70 diagnostics or even therapeutics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes.Methods. 2016 Mar 15;97:51-7. doi: 10.1016/j.ymeth.2015.12.005. Epub 2015 Dec 8. Methods. 2016. PMID: 26678795
-
Directed Evolution of Aptamer Discovery Technologies.Acc Chem Res. 2022 Mar 1;55(5):685-695. doi: 10.1021/acs.accounts.1c00724. Epub 2022 Feb 7. Acc Chem Res. 2022. PMID: 35130439
-
Advancements in Aptamer Discovery Technologies.Acc Chem Res. 2016 Sep 20;49(9):1903-10. doi: 10.1021/acs.accounts.6b00283. Epub 2016 Aug 15. Acc Chem Res. 2016. PMID: 27526193
-
In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies.RNA Biol. 2016 Dec;13(12):1232-1245. doi: 10.1080/15476286.2016.1236173. Epub 2016 Oct 7. RNA Biol. 2016. PMID: 27715478 Free PMC article. Review.
-
Methods for Improving Aptamer Binding Affinity.Molecules. 2016 Mar 28;21(4):421. doi: 10.3390/molecules21040421. Molecules. 2016. PMID: 27043498 Free PMC article. Review.
Cited by
-
Zwitterionic DNA: enzymatic synthesis of hypermodified DNA bearing four different cationic substituents at all four nucleobases.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf155. doi: 10.1093/nar/gkaf155. Nucleic Acids Res. 2025. PMID: 40057376 Free PMC article.
-
Superanionic DNA: enzymatic synthesis of hypermodified DNA bearing four different anionic substituents at all four nucleobases.Nucleic Acids Res. 2023 Nov 27;51(21):11428-11438. doi: 10.1093/nar/gkad893. Nucleic Acids Res. 2023. PMID: 37870471 Free PMC article.
-
Selection of optimised ligands by fluorescence-activated bead sorting.Chem Sci. 2023 Aug 11;14(35):9517-9525. doi: 10.1039/d3sc03581f. eCollection 2023 Sep 13. Chem Sci. 2023. PMID: 37712023 Free PMC article.
-
Arylethynyl- or Alkynyl-Linked Pyrimidine and 7-Deazapurine 2'-Deoxyribonucleoside 3'-Phosphoramidites for Chemical Synthesis of Hypermodified Hydrophobic Oligonucleotides.ACS Omega. 2023 Oct 12;8(42):39447-39453. doi: 10.1021/acsomega.3c05202. eCollection 2023 Oct 24. ACS Omega. 2023. PMID: 37901526 Free PMC article.
-
Strict Interactions of Fifth Letters, Hydrophobic Unnatural Bases, in XenoAptamers with Target Proteins.J Am Chem Soc. 2023 Sep 20;145(37):20432-20441. doi: 10.1021/jacs.3c06122. Epub 2023 Sep 7. J Am Chem Soc. 2023. PMID: 37677157 Free PMC article.
References
LinkOut - more resources
Full Text Sources

