Prognostic values of tissue-resident CD8+T cells in human hepatocellular carcinoma and intrahepatic cholangiocarcinoma
- PMID: 37024870
- PMCID: PMC10077621
- DOI: 10.1186/s12957-023-03009-6
Prognostic values of tissue-resident CD8+T cells in human hepatocellular carcinoma and intrahepatic cholangiocarcinoma
Abstract
Background: Tissue-resident CD8+T cells (CD103+CD8+T cells) are the essential effector cell population of anti-tumor immune response in tissue regional immunity. And we have reported that IL-33 can promote the proliferation and effector function of tissue-resident CD103+CD8+T cells. As of now, the immunolocalization and the prognostic values of tissue-resident CD8+T cells in human hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) still remain to be illustrated.
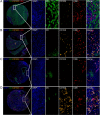
Methods: In our present study, we used the tissue microarrays of HCC and ICC, the multicolor immunohistochemistry (mIHC), and imaging analysis to characterize the tissue-resident CD8+T cells in HCC and ICC tissues. The prognostic values and clinical associations were also analyzed. We also studied the biological functions and the cell-cell communication between tumor-infiltrating CD103+CD8+T cells and other cell types in HCC and ICC based on the published single-cell RNA sequencing (scRNA-seq) data.
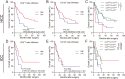
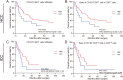
Results: Our work unveiled the expressions of CD8 and CD103 and immunolocalization of tissue-resident CD8+T cells in human HCC and ICC. Elevated CD8+T cells indicated a better overall survival (OS) rate, implying that tumor-infiltrating CD8+T cells in HCC and ICC could serve as an independent prognostic factor. Moreover, the number of CD103+CD8+T cells was increased in HCC and ICC tissues compared with adjacent normal tissues. HCC patients defined as CD8highCD103high had a better OS, and the CD8lowCD103low group tended to have a poorer prognosis in ICC. Evaluation of the CD103+CD8+T-cell ratio in CD8+T cells could also be a prognostic predictor for HCC and ICC patients. A higher ratio of CD103+CD8+T cells over total CD8+T cells in HCC tissues was negatively and significantly associated with the advanced pathological stage. The percentage of higher numbers of CD103+CD8+T cells in ICC tissues was negatively and significantly associated with the advanced pathological stage. In contrast, the higher ratio of CD103+CD8+T cells over total CD8+T cells in ICC tissues was negatively and significantly associated with the advanced pathological stage. In addition, single-cell transcriptomics revealed that CD103+CD8+T cells were enriched in genes associated with T-cell activation, proliferation, cytokine function, and T-cell exhaustion.
Conclusion: The CD103+ tumor-specific T cells signified an important prognostic marker with improved OS, and the evaluation of the tissue-resident CD103+CD8+T cells might be helpful in assessing the on-treatment response of liver cancer.
Keywords: Hepatocellular carcinoma; Intrahepatic Cholangiocarcinoma; Multicolor immunohistochemistry; Prognosis; Tissue-resident CD103+CD8+T cells.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Xue R, Chen L, Zhang C, Fujita M, Li R, Yan SM, Ong CK, Liao X, Gao Q, Sasagawa S, et al. Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell. 2019;35(6):932–947.e938. doi: 10.1016/j.ccell.2019.04.007. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

