Identification of cell barcodes from long-read single-cell RNA-seq with BLAZE
- PMID: 37024980
- PMCID: PMC10077662
- DOI: 10.1186/s13059-023-02907-y
Identification of cell barcodes from long-read single-cell RNA-seq with BLAZE
Abstract
Long-read single-cell RNA sequencing (scRNA-seq) enables the quantification of RNA isoforms in individual cells. However, long-read scRNA-seq using the Oxford Nanopore platform has largely relied upon matched short-read data to identify cell barcodes. We introduce BLAZE, which accurately and efficiently identifies 10x cell barcodes using only nanopore long-read scRNA-seq data. BLAZE outperforms the existing tools and provides an accurate representation of the cells present in long-read scRNA-seq when compared to matched short reads. BLAZE simplifies long-read scRNA-seq while improving the results, is compatible with downstream tools accepting a cell barcode file, and is available at https://github.com/shimlab/BLAZE .
© 2023. The Author(s).
Conflict of interest statement
Y.Y, Y.D.P, R.D.P, and M.B.C have received support from Oxford Nanopore Technologies (ONT) to present their findings at scientific conferences. However, ONT played no role in the study design, execution, analysis, or publication.
Figures





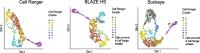
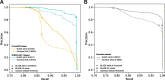
References
-
- Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, Chen H, Wang J, Tang H, Ge W, et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. - PubMed
-
- Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks G-J, Larsson AJM, Faridani OR, Sandberg R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38:708–714. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

