Genome-wide characterization and expression analysis of the MLO gene family sheds light on powdery mildew resistance in Lagenaria siceraria
- PMID: 37025859
- PMCID: PMC10070393
- DOI: 10.1016/j.heliyon.2023.e14624
Genome-wide characterization and expression analysis of the MLO gene family sheds light on powdery mildew resistance in Lagenaria siceraria
Abstract
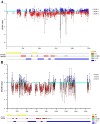
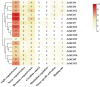
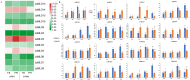
MLO (mildew locus O) genes play a vital role in plant disease defense system, especially powdery mildew (PM). Lagenaria siceraria is a distinct Cucurbitaceae crop, and PM is one of the most serious diseases threatening crop production and quality. Although MLOs have been exploited in many Cucurbitaceae species, genome-wide mining of MLO gene family in bottle gourd has not been surveyed yet. Here we identified 16 MLO genes in our recently assembled L. siceraria genome. A total of 343 unique MLO protein sequences from 20 species were characterized and compared to deduce a generally high level of purifying selection and the occurrence of regions related to candidate susceptibility factors in the evolutional divergence. LsMLOs were clustered in six clades containing seven conserved transmembrane domains and 10 clade-specific motifs along with deletion and variation. Three genes (LsMLO3, LsMLO6, and LsMLO13) in clade V showed high sequence identity with orthologues involved in PM susceptibility. The expression pattern of LsMLOs was tissue-specific but not cultivar-specific. Furthermore, it was indicated by qRT-PCR and RNA-seq that LsMLO3 and LsMLO13 were highly upregulated in response to PM stress. Subsequent sequence analysis revealed the structural deletion of LsMLO13 and a single nonsynonymous substitution of LsMLO3 in the PM-resistant genotype. Taken all together, it is speculated that LsMLO13 is likely a major PM susceptibility factor. The results of this study provide new insights into MLO family genes in bottle gourd and find a potential candidate S gene for PM tolerance breeding.
Keywords: Bottle gourd; Expression analysis; MLO family; Phylogenetic analysis; Powdery mildew.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The First Genome-Wide Mildew Locus O Genes Characterization in the Lamiaceae Plant Family.Int J Mol Sci. 2023 Sep 4;24(17):13627. doi: 10.3390/ijms241713627. Int J Mol Sci. 2023. PMID: 37686433 Free PMC article.
-
Structure, evolution and functional inference on the Mildew Locus O (MLO) gene family in three cultivated Cucurbitaceae spp.BMC Genomics. 2015 Dec 29;16:1112. doi: 10.1186/s12864-015-2325-3. BMC Genomics. 2015. PMID: 26715041 Free PMC article.
-
Candidate gene mapping reveals VrMLO12 (MLO Clade II) is associated with powdery mildew resistance in mungbean (Vigna radiata [L.] Wilczek).Plant Sci. 2020 Sep;298:110594. doi: 10.1016/j.plantsci.2020.110594. Epub 2020 Jul 6. Plant Sci. 2020. PMID: 32771151
-
Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol. 2014 Oct;204(2):273-81. doi: 10.1111/nph.12889. New Phytol. 2014. PMID: 25453131 Review.
-
Unmasking Mildew Resistance Locus O.Trends Plant Sci. 2021 Oct;26(10):1006-1013. doi: 10.1016/j.tplants.2021.05.009. Epub 2021 Jun 23. Trends Plant Sci. 2021. PMID: 34175219 Review.
Cited by
-
Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations.Plants (Basel). 2025 Jul 8;14(14):2091. doi: 10.3390/plants14142091. Plants (Basel). 2025. PMID: 40733328 Free PMC article. Review.
-
Comparative Genome Analysis of Old World and New World TYLCV Reveals a Biasness toward Highly Variable Amino Acids in Coat Protein.Plants (Basel). 2023 May 16;12(10):1995. doi: 10.3390/plants12101995. Plants (Basel). 2023. PMID: 37653912 Free PMC article.
References
LinkOut - more resources
Full Text Sources

