Distinct regulatory functions and biological roles of lncRNA splice variants
- PMID: 37025931
- PMCID: PMC10070373
- DOI: 10.1016/j.omtn.2023.03.004
Distinct regulatory functions and biological roles of lncRNA splice variants
Abstract
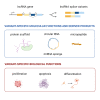
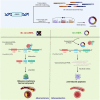
Alternative splicing (AS) of RNA molecules is a key contributor to transcriptome diversity. In humans, 90%-95% of multi-exon genes produce alternatively spliced RNA transcripts. Therefore, every single gene has the opportunity of producing multiple splice variants, including long non-coding RNA (lncRNA) genes that undergo RNA maturation steps such as conventional and alternative splicing. Emerging evidence suggests significant roles for these lncRNA splice variants in many aspects of cell biology. Differential changes in expression of specific lncRNA splice variants have also been associated with many diseases including cancer. This review covers the current knowledge on this emerging topic of investigation. We provide exclusive insights on the AS landscape of lncRNAs and also describe at the molecular level the functional relevance of lncRNA splice variants, i.e., RNA-based differential functions, production of micropeptides, and generation of circular RNAs. Finally, we discuss exciting perspectives for this emerging field and outline the work required to further develop research endeavors in this field.
Keywords: MT: Non-coding RNAs; alternative splicing; circular RNA; long non-coding RNA; micropeptide; splice variant; therapeutics.
© 2023 The Author(s).
Conflict of interest statement
The authors have declared no competing interest.
Figures





References
-
- Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. - PubMed
-
- Irimia M., Blencowe B.J. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 2012;24:323–332. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

