Emergence and circulation of azole-resistant C. albicans, C. auris and C. parapsilosis bloodstream isolates carrying Y132F, K143R or T220L Erg11p substitutions in Colombia
- PMID: 37026059
- PMCID: PMC10070958
- DOI: 10.3389/fcimb.2023.1136217
Emergence and circulation of azole-resistant C. albicans, C. auris and C. parapsilosis bloodstream isolates carrying Y132F, K143R or T220L Erg11p substitutions in Colombia
Abstract
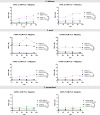
Methods: Over a four-year period, 123 Candida bloodstream isolates were collected at a quaternary care hospital. The isolates were identified by MALDI-TOF MS and their fluconazole (FLC) susceptibility patterns were assessed according to CLSI guidelines. Subsequently, sequencing of ERG11, TAC1 or MRR1, and efflux pump activity were performed for resistant isolates.
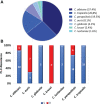
Results: Out of 123 clinical strains,C. albicans accounted for 37.4%, followed by C. tropicalis 26.8%, C. parapsilosis 19.5%, C. auris 8.1%, C. glabrata 4.1%, C. krusei 2.4% and C. lusitaniae 1.6%. Resistance to FLC reached 18%; in addition, a high proportion of isolates were cross-resistant to voriconazole. Erg11 amino acid substitutions associated with FLC-resistance (Y132F, K143R, or T220L) were found in 11/19 (58%) of FLCresistant isolates. Furthermore, novel mutations were found in all genes evaluated. Regarding efflux pumps, 8/19 (42%) of FLC-resistant Candida spp strains showed significant efflux activity. Finally, 6/19 (31%) of FLC-resistant isolates neither harbored resistance-associated mutations nor showed efflux pump activity. Among FLC-resistant species, C. auris 7/10 (70%) and C. parapsilosis 6/24 (25%) displayed the highest percentages of resistance (C. albicans 6/46, 13%).
Discussion: Overall, 68% of FLC-resistant isolates exhibited a mechanism that could explain their phenotype (e.g. mutations, efflux pump activity, or both). We provide evidence that isolates from patients admitted to a Colombian hospital harbor amino acid substitutions related to resistance to one of the most commonly used molecules in the hospital setting, with Y132F being the most frequently detected.
Keywords: Candida species; Colombia; ERG11; Y132F; bloodstream infections; fluconazole resistance.
Copyright © 2023 Ceballos-Garzon, Peñuela, Valderrama-Beltrán, Vargas-Casanova, Ariza and Parra-Giraldo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Arastehfar A., Daneshnia F., Hilmioglu-Polat S., Fang W., Yaşar M., Polat F., et al. (2020). First report of candidemia clonal outbreak caused by emerging fluconazole-resistant candida parapsilosis isolates harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob. Agents Chemother. 64 , e01001-20. doi: 10.1128/AAC.01001-20/SUPPL_FILE/AAC.01001-20-S0001.PDF - DOI - PMC - PubMed
-
- Berkow E. L., Manigaba K., Parker J. E., Barker K. S., Kelly S. L., Rogers P. D. (2015). Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in candida parapsilosis. Antimicrob. Agents Chemother. 59, 5942–5950. doi: 10.1128/AAC.01358-15 - DOI - PMC - PubMed
-
- Branco J., Silva A. P., Silva R. M., Silva-Dias A., Pina-Vaz C., Butler G., et al. (2015). Fluconazole and voriconazole resistance in candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob. Agents Chemother. 59 , 6629-33. doi: 10.1128/AAC.00842-15 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

