The ADAR1 editome reveals drivers of editing-specificity for ADAR1-isoforms
- PMID: 37026479
- PMCID: PMC10201426
- DOI: 10.1093/nar/gkad265
The ADAR1 editome reveals drivers of editing-specificity for ADAR1-isoforms
Abstract
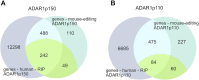
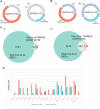
Adenosine deaminase acting on RNA ADAR1 promotes A-to-I conversion in double-stranded and structured RNAs. ADAR1 has two isoforms transcribed from different promoters: cytoplasmic ADAR1p150 is interferon-inducible while ADAR1p110 is constitutively expressed and primarily localized in the nucleus. Mutations in ADAR1 cause Aicardi - Goutières syndrome (AGS), a severe autoinflammatory disease associated with aberrant IFN production. In mice, deletion of ADAR1 or the p150 isoform leads to embryonic lethality driven by overexpression of interferon-stimulated genes. This phenotype is rescued by deletion of the cytoplasmic dsRNA-sensor MDA5 indicating that the p150 isoform is indispensable and cannot be rescued by ADAR1p110. Nevertheless, editing sites uniquely targeted by ADAR1p150 remain elusive. Here, by transfection of ADAR1 isoforms into ADAR-less mouse cells we detect isoform-specific editing patterns. Using mutated ADAR variants, we test how intracellular localization and the presence of a Z-DNA binding domain-α affect editing preferences. These data show that ZBDα only minimally contributes to p150 editing-specificity while isoform-specific editing is primarily directed by the intracellular localization of ADAR1 isoforms. Our study is complemented by RIP-seq on human cells ectopically expressing tagged-ADAR1 isoforms. Both datasets reveal enrichment of intronic editing and binding by ADAR1p110 while ADAR1p150 preferentially binds and edits 3'UTRs.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

