Establishment and characterization of a novel cancer stem-like cell of cholangiocarcinoma
- PMID: 37026527
- PMCID: PMC10394157
- DOI: 10.1111/cas.15812
Establishment and characterization of a novel cancer stem-like cell of cholangiocarcinoma
Abstract
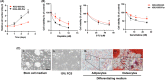
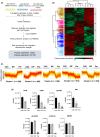
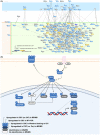
Cholangiocarcinoma (CCA) is an aggressive malignant tumor of bile duct epithelia. Recent evidence suggests the impact of cancer stem cells (CSC) on the therapeutic resistance of CCA; however, the knowledge of CSC in CCA is limited due to the lack of a CSC model. In this study, we successfully established a stable sphere-forming CCA stem-like cell, KKU-055-CSC, from the original CCA cell line, KKU-055. The KKU-055-CSC exhibits CSC characteristics, including: (1) the ability to grow stably and withstand continuous passage for a long period of culture in the stem cell medium, (2) high expression of stem cell markers, (3) low responsiveness to standard chemotherapy drugs, (4) multilineage differentiation, and (5) faster and constant expansive tumor formation in xenograft mouse models. To identify the CCA-CSC-associated pathway, we have undertaken a global proteomics and functional cluster/network analysis. Proteomics identified the 5925 proteins in total, and the significantly upregulated proteins in CSC compared with FCS-induced differentiated CSC and its parental cells were extracted. Network analysis revealed that high mobility group A1 (HMGA1) and Aurora A signaling through the signal transducer and activator of transcription 3 pathways were enriched in KKU-055-CSC. Knockdown of HMGA1 in KKU-055-CSC suppressed the expression of stem cell markers, induced the differentiation followed by cell proliferation, and enhanced sensitivity to chemotherapy drugs including Aurora A inhibitors. In silico analysis indicated that the expression of HMGA1 was correlated with Aurora A expressions and poor survival of CCA patients. In conclusion, we have established a unique CCA stem-like cell model and identified the HMGA1-Aurora A signaling as an important pathway for CSC-CCA.
Keywords: HMGA1; bile duct; cancer stem cell; cholangiocarcinoma; liver.
© 2023 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Yoshihiro Komohara is currently an Editorial Board Member of
Figures






References
-
- Lee KJ, Yi SW, Cha J, et al. A pilot study of concurrent chemoradiotherapy with gemcitabine and cisplatin in patients with locally advanced biliary tract cancer. Cancer Chemother Pharmacol. 2016;78:841‐846. - PubMed
-
- Sripa B, Seubwai W, Vaeteewoottacharn K, et al. Functional and genetic characterization of three cell lines derived from a single tumor of an Opisthorchis viverrini‐associated cholangiocarcinoma patient. Hum Cell. 2020;33:695‐708. - PubMed
-
- Kawamoto M, Umebayashi M, Tanaka H, et al. Combined gemcitabine and metronidazole is a promising therapeutic strategy for cancer stem‐like cholangiocarcinoma. Anticancer Res. 2018;38:2739‐2748. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

