BICePs v2.0: Software for Ensemble Reweighting Using Bayesian Inference of Conformational Populations
- PMID: 37027181
- PMCID: PMC10278562
- DOI: 10.1021/acs.jcim.2c01296
BICePs v2.0: Software for Ensemble Reweighting Using Bayesian Inference of Conformational Populations
Abstract
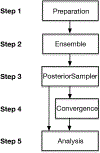
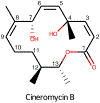
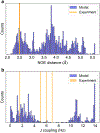
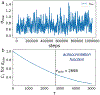
Bayesian Inference of Conformational Populations (BICePs) version 2.0 (v2.0) is a free, open-source Python package that reweights theoretical predictions of conformational state populations using sparse and/or noisy experimental measurements. In this article, we describe the implementation and usage of the latest version of BICePs (v2.0), a powerful, user-friendly and extensible package which makes several improvements upon the previous version. The algorithm now supports many experimental NMR observables (NOE distances, chemical shifts, J-coupling constants, and hydrogen-deuterium exchange protection factors), and enables convenient data preparation and processing. BICePs v2.0 can perform automatic analysis of the sampled posterior, including visualization, and evaluation of statistical significance and sampling convergence. We provide specific coding examples for these topics, and present a detailed example illustrating how to use BICePs v2.0 to reweight a theoretical ensemble using experimental measurements.
Figures

Similar articles
-
Model Selection Using BICePs: A Bayesian Approach for Force Field Validation and Parameterization.J Phys Chem B. 2018 May 31;122(21):5610-5622. doi: 10.1021/acs.jpcb.7b11871. Epub 2018 Mar 23. J Phys Chem B. 2018. PMID: 29518328 Free PMC article.
-
Reconciling Simulated Ensembles of Apomyoglobin with Experimental Hydrogen/Deuterium Exchange Data Using Bayesian Inference and Multiensemble Markov State Models.J Chem Theory Comput. 2020 Feb 11;16(2):1333-1348. doi: 10.1021/acs.jctc.9b01240. Epub 2020 Jan 23. J Chem Theory Comput. 2020. PMID: 31917926
-
Bayesian inference of conformational state populations from computational models and sparse experimental observables.J Comput Chem. 2014 Nov 15;35(30):2215-24. doi: 10.1002/jcc.23738. Epub 2014 Sep 24. J Comput Chem. 2014. PMID: 25250719
-
Reconciling Simulations and Experiments With BICePs: A Review.Front Mol Biosci. 2021 May 11;8:661520. doi: 10.3389/fmolb.2021.661520. eCollection 2021. Front Mol Biosci. 2021. PMID: 34046431 Free PMC article. Review.
-
A primer on Bayesian inference for biophysical systems.Biophys J. 2015 May 5;108(9):2103-13. doi: 10.1016/j.bpj.2015.03.042. Biophys J. 2015. PMID: 25954869 Free PMC article. Review.
Cited by
-
MELD in Action: Harnessing Data to Accelerate Molecular Dynamics.J Chem Inf Model. 2025 Feb 24;65(4):1685-1693. doi: 10.1021/acs.jcim.4c02108. Epub 2025 Feb 2. J Chem Inf Model. 2025. PMID: 39893583 Review.
-
Computational Methods to Investigate Intrinsically Disordered Proteins and their Complexes.ArXiv [Preprint]. 2024 Sep 3:arXiv:2409.02240v1. ArXiv. 2024. PMID: 39279844 Free PMC article. Preprint.
-
Effects of All-Atom and Coarse-Grained Molecular Mechanics Force Fields on Amyloid Peptide Assembly: The Case of a Tau K18 Monomer.J Chem Inf Model. 2024 Dec 9;64(23):8880-8891. doi: 10.1021/acs.jcim.4c01448. Epub 2024 Nov 23. J Chem Inf Model. 2024. PMID: 39579121 Free PMC article.
-
Automatic Forward Model Parameterization with Bayesian Inference of Conformational Populations.ArXiv [Preprint]. 2025 Jun 18:arXiv:2405.18532v2. ArXiv. 2025. PMID: 38855540 Free PMC article. Preprint.
-
Combining simulations and experiments - a perspective on maximum entropy methods.Phys Chem Chem Phys. 2025 Jul 17;27(28):14704-14717. doi: 10.1039/d5cp01263e. Phys Chem Chem Phys. 2025. PMID: 40600335 Free PMC article. Review.
References
-
- Rieping W; Habeck M; Nilges M Inferential Structure Determination. Science 2005, 309, 303–306. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

