Minimal Out-of-Equilibrium Metabolism for Synthetic Cells: A Membrane Perspective
- PMID: 37027340
- PMCID: PMC10127287
- DOI: 10.1021/acssynbio.3c00062
Minimal Out-of-Equilibrium Metabolism for Synthetic Cells: A Membrane Perspective
Abstract
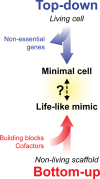
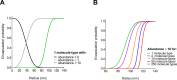
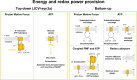
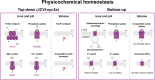
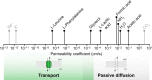
Life-like systems need to maintain a basal metabolism, which includes importing a variety of building blocks required for macromolecule synthesis, exporting dead-end products, and recycling cofactors and metabolic intermediates, while maintaining steady internal physical and chemical conditions (physicochemical homeostasis). A compartment, such as a unilamellar vesicle, functionalized with membrane-embedded transport proteins and metabolic enzymes encapsulated in the lumen meets these requirements. Here, we identify four modules designed for a minimal metabolism in a synthetic cell with a lipid bilayer boundary: energy provision and conversion, physicochemical homeostasis, metabolite transport, and membrane expansion. We review design strategies that can be used to fulfill these functions with a focus on the lipid and membrane protein composition of a cell. We compare our bottom-up design with the equivalent essential modules of JCVI-syn3a, a top-down genome-minimized living cell with a size comparable to that of large unilamellar vesicles. Finally, we discuss the bottlenecks related to the insertion of a complex mixture of membrane proteins into lipid bilayers and provide a semiquantitative estimate of the relative surface area and lipid-to-protein mass ratios (i.e., the minimal number of membrane proteins) that are required for the construction of a synthetic cell.
Keywords: JCVI-syn3a; bottom-up synthetic cells; energy conservation; membrane composition; metabolite transport; minimal metabolism; out-of-equilibrium; physicochemical homeostasis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Breuer M.; Earnest T. M; Merryman C.; Wise K. S; Sun L.; Lynott M. R; Hutchison C. A; Smith H. O; Lapek J. D; Gonzalez D. J; de Crecy-Lagard V.; Haas D.; Hanson A. D; Labhsetwar P.; Glass J. I; Luthey-Schulten Z. Essential Metabolism for a Minimal Cell. Elife 2019, 8, e36842.10.7554/eLife.36842. - DOI - PMC - PubMed
-
- Reuß D. R.; Altenbuchner J.; Mäder U.; Rath H.; Ischebeck T.; Sappa P. K.; Thürmer A.; Guérin C.; Nicolas P.; Steil L.; Zhu B.; Feussner I.; Klumpp S.; Daniel R.; Commichau F. M.; Völker U.; Stülke J. Large-Scale Reduction of the Bacillus Subtilis Genome: Consequences for the Transcriptional Network, Resource Allocation, and Metabolism. Genome Res. 2017, 27 (2), 289–299. 10.1101/gr.215293.116. - DOI - PMC - PubMed
-
- Hashimoto M.; Ichimura T.; Mizoguchi H.; Tanaka K.; Fujimitsu K.; Keyamura K.; Ote T.; Yamakawa T.; Yamazaki Y.; Mori H.; Katayama T.; Kato J. I. Cell Size and Nucleoid Organization of Engineered Escherichia Coli Cells with a Reduced Genome. Mol. Microbiol. 2005, 55 (1), 137–149. 10.1111/j.1365-2958.2004.04386.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

