Eosinophils promote effector functions of lung group 2 innate lymphoid cells in allergic airway inflammation in mice
- PMID: 37028525
- PMCID: PMC10503660
- DOI: 10.1016/j.jaci.2023.03.023
Eosinophils promote effector functions of lung group 2 innate lymphoid cells in allergic airway inflammation in mice
Abstract
Background: Group 2 innate lymphoid cells (ILC2s) are critical mediators of type 2 respiratory inflammation, releasing IL-5 and IL-13 and promoting the pulmonary eosinophilia associated with allergen provocation. Although ILC2s have been shown to promote eosinophil activities, the role of eosinophils in group 2 innate lymphoid cell (ILC2) responses is less well defined.
Objective: We sought to investigate the role of eosinophils in activation of ILC2s in models of allergic asthma and in vitro.
Methods: Inducible eosinophil-deficient mice were exposed to allergic respiratory inflammation models of asthma, such as ovalbumin or house dust mite challenge, or to innate models of type 2 airway inflammation, such as inhalation of IL-33. Eosinophil-specific IL-4/13-deficient mice were used to address the specific roles for eosinophil-derived cytokines. Direct cell interactions between ILC2s and eosinophils were assessed by in vitro culture experiments.
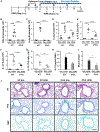
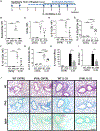
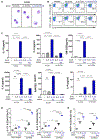
Results: Targeted depletion of eosinophils resulted in significant reductions of total and IL-5+ and IL-13+ lung ILC2s in all models of respiratory inflammation. This correlated with reductions in IL-13 levels and mucus in the airway. Eosinophil-derived IL-4/13 was necessary for both eosinophil and ILC2 accumulation in lung in allergen models. In vitro, eosinophils released soluble mediators that induced ILC2 proliferation and G protein-coupled receptor-dependent chemotaxis of ILC2s. Coculture of ILC2s and IL-33-activated eosinophils resulted in transcriptome changes in both ILC2s and eosinophils, suggesting potential novel reciprocal interactions.
Conclusion: These studies demonstrate that eosinophils play a reciprocal role in ILC2 effector functions as part of both adaptive and innate type 2 pulmonary inflammatory events.
Keywords: Eosinophil; IL-13; IL-33; IL-4; asthma; eosinophil deficient; group 2 innate lymphoid cell; inflammation; lung.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Figures


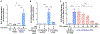


Similar articles
-
MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation.J Allergy Clin Immunol. 2017 Mar;139(3):1007-1016.e9. doi: 10.1016/j.jaci.2016.06.035. Epub 2016 Aug 1. J Allergy Clin Immunol. 2017. PMID: 27492144
-
Protein kinase Cθ controls type 2 innate lymphoid cell and TH2 responses to house dust mite allergen.J Allergy Clin Immunol. 2017 May;139(5):1650-1666. doi: 10.1016/j.jaci.2016.08.044. Epub 2016 Oct 14. J Allergy Clin Immunol. 2017. PMID: 27746240
-
Interplay Between the IL-33/ST2 Axis and Bone Marrow ILC2s in Protease Allergen-Induced IL-5-Dependent Eosinophilia.Front Immunol. 2020 Jun 2;11:1058. doi: 10.3389/fimmu.2020.01058. eCollection 2020. Front Immunol. 2020. PMID: 32582171 Free PMC article.
-
Group 2 innate lymphoid cells (ILC2s): The spotlight in asthma pathogenesis and lung tissue injury.Allergol Immunopathol (Madr). 2021 Mar 1;49(2):208-216. doi: 10.15586/aei.v49i2.29. eCollection 2021. Allergol Immunopathol (Madr). 2021. PMID: 33641310 Review.
-
Type 2 innate lymphoid cells: at the cross-roads in allergic asthma.Semin Immunopathol. 2016 Jul;38(4):483-96. doi: 10.1007/s00281-016-0556-2. Epub 2016 Mar 10. Semin Immunopathol. 2016. PMID: 26965110 Free PMC article. Review.
Cited by
-
A deep insight into ferroptosis in lung disease: facts and perspectives.Front Oncol. 2024 Mar 18;14:1354859. doi: 10.3389/fonc.2024.1354859. eCollection 2024. Front Oncol. 2024. PMID: 38562175 Free PMC article. Review.
-
Integrative Cross-Talk in Asthma: Unraveling the Complex Interactions Between Eosinophils, Immune, and Structural Cells in the Airway Microenvironment.Diagnostics (Basel). 2024 Oct 31;14(21):2448. doi: 10.3390/diagnostics14212448. Diagnostics (Basel). 2024. PMID: 39518415 Free PMC article. Review.
-
The immunology of asthma and chronic rhinosinusitis.Nat Rev Immunol. 2025 Aug;25(8):569-587. doi: 10.1038/s41577-025-01159-0. Epub 2025 Apr 16. Nat Rev Immunol. 2025. PMID: 40240657 Review.
-
Zinc Deficiency and Zinc Supplementation in Allergic Diseases.Biomolecules. 2024 Jul 19;14(7):863. doi: 10.3390/biom14070863. Biomolecules. 2024. PMID: 39062576 Free PMC article. Review.
-
Depletion of eosinophils during sensitization but not challenge phase in mice blocks the development of food allergy early in life.J Immunol. 2025 Apr 1;214(4):582-594. doi: 10.1093/jimmun/vkae044. J Immunol. 2025. PMID: 40073088
References
-
- Hammad H, Lambrecht BN. The basic immunology of asthma. Cell 2021;184:1469–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

