VIRMA promotes nasopharyngeal carcinoma, tumorigenesis, and metastasis by upregulation of E2F7 in an m6A-dependent manner
- PMID: 37028765
- PMCID: PMC10235437
- DOI: 10.1016/j.jbc.2023.104677
VIRMA promotes nasopharyngeal carcinoma, tumorigenesis, and metastasis by upregulation of E2F7 in an m6A-dependent manner
Abstract
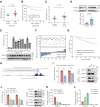
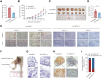
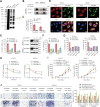
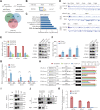
The N6-methyladenosine (m6A) modification possesses new and essential roles in tumor initiation and progression by regulating mRNA biology. However, the role of aberrant m6A regulation in nasopharyngeal carcinoma (NPC) remains unclear. Here, through comprehensive analyses of NPC cohorts from the GEO database and our internal cohort, we identified that VIRMA, an m6A writer, is significantly upregulated in NPC and plays an essential role in tumorigenesis and metastasis of NPC, both in vitro and in vivo. High VIRMA expression served as a prognostic biomarker and was associated with poor outcomes in patients with NPC. Mechanistically, VIRMA mediated the m6A methylation of E2F7 3'-UTR, then IGF2BP2 bound, and maintained the stability of E2F7 mRNA. An integrative high-throughput sequencing approach revealed that E2F7 drives a unique transcriptome distinct from the classical E2F family in NPC, which functioned as an oncogenic transcriptional activator. E2F7 cooperated with CBFB-recruited RUNX1 in a non-canonical manner to transactivate ITGA2, ITGA5, and NTRK1, strengthening Akt signaling-induced tumor-promoting effect.
Keywords: E2F7; VIRMA; m6A; metastasis; nasopharyngeal carcinoma.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
m6A-enriched lncRNA LINC00839 promotes tumor progression by enhancing TAF15-mediated transcription of amine oxidase AOC1 in nasopharyngeal carcinoma.J Biol Chem. 2023 Jul;299(7):104873. doi: 10.1016/j.jbc.2023.104873. Epub 2023 May 29. J Biol Chem. 2023. PMID: 37257820 Free PMC article.
-
HNRNPH1 stabilizes FLOT2 mRNA in a non-canonical m6A-dependent manner to promote malignant progression in nasopharyngeal carcinoma.Cell Oncol (Dordr). 2024 Dec;47(6):2279-2295. doi: 10.1007/s13402-024-01016-7. Epub 2024 Nov 21. Cell Oncol (Dordr). 2024. PMID: 39570559
-
Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3.Cancer Res. 2019 Sep 15;79(18):4612-4626. doi: 10.1158/0008-5472.CAN-19-0799. Epub 2019 Jul 22. Cancer Res. 2019. PMID: 31331909
-
Role of m6A methyltransferase component VIRMA in multiple human cancers (Review).Cancer Cell Int. 2021 Mar 17;21(1):172. doi: 10.1186/s12935-021-01868-1. Cancer Cell Int. 2021. PMID: 33731118 Free PMC article. Review.
-
Role of N6-methyladenosine methylation in nasopharyngeal carcinoma: current insights and future prospective.Cell Death Discov. 2024 Dec 18;10(1):490. doi: 10.1038/s41420-024-02266-y. Cell Death Discov. 2024. PMID: 39695216 Free PMC article. Review.
Cited by
-
The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy.Biomolecules. 2025 Jun 22;15(7):912. doi: 10.3390/biom15070912. Biomolecules. 2025. PMID: 40723784 Free PMC article. Review.
-
Hyperactivation of mTOR/eIF4E Signaling Pathway Promotes the Production of Tryptophan-To-Phenylalanine Substitutants in EBV-Positive Gastric Cancer.Adv Sci (Weinh). 2024 Sep;11(35):e2402284. doi: 10.1002/advs.202402284. Epub 2024 Jul 12. Adv Sci (Weinh). 2024. PMID: 38994917 Free PMC article.
-
The m6A demethylases FTO and ALKBH5 aggravate the malignant progression of nasopharyngeal carcinoma by coregulating ARHGAP35.Cell Death Discov. 2024 Jan 23;10(1):43. doi: 10.1038/s41420-024-01810-0. Cell Death Discov. 2024. PMID: 38263362 Free PMC article.
-
Research Progress of Liquid Biopsy Based on DNA Methylation in Tumor Diagnosis and Treatment.Biomolecules. 2024 Dec 19;14(12):1634. doi: 10.3390/biom14121634. Biomolecules. 2024. PMID: 39766341 Free PMC article. Review.
-
Liquid-liquid phase separation in diseases.MedComm (2020). 2024 Jul 13;5(7):e640. doi: 10.1002/mco2.640. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 39006762 Free PMC article. Review.
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Chen Y.P., Chan A.T.C., Le Q.T., Blanchard P., Sun Y., Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. - PubMed
-
- Dawson M.A., Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

