A duplex tetra-primer ARMS-PCR assay to discriminate three species of the Schistosoma haematobium group: Schistosoma curassoni, S. bovis, S. haematobium and their hybrids
- PMID: 37029440
- PMCID: PMC10082484
- DOI: 10.1186/s13071-023-05754-9
A duplex tetra-primer ARMS-PCR assay to discriminate three species of the Schistosoma haematobium group: Schistosoma curassoni, S. bovis, S. haematobium and their hybrids
Abstract
Background: The use of applications involving single nucleotide polymorphisms (SNPs) has greatly increased since the beginning of the 2000s, with the number of associated techniques expanding rapidly in the field of molecular research. Tetra-primer amplification refractory mutation system-PCR (T-ARMS-PCR) is one such technique involving SNP genotyping. It has the advantage of amplifying multiple alleles in a single reaction with the inclusion of an internal molecular control. We report here the development of a rapid, reliable and cost-effective duplex T-ARMS-PCR assay to distinguish between three Schistosoma species, namely Schistosoma haematobium (human parasite), Schistosoma bovis and Schistosoma curassoni (animal parasites), and their hybrids. This technique will facilitate studies of population genetics and the evolution of introgression events.
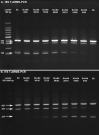
Methods: During the development of the technique we focused on one of the five inter-species internal transcribed spacer (ITS) SNPs and one of the inter-species 18S SNPs which, when combined, discriminate between all three Schistosoma species and their hybrid forms. We designed T-ARMS-PCR primers to amplify amplicons of specific lengths for each species, which in turn can then be visualized on an electrophoresis gel. This was further tested using laboratory and field-collected adult worms and field-collected larval stages (miracidia) from Spain, Egypt, Mali, Senegal and Ivory Coast. The combined duplex T-ARMS-PCR and ITS + 18S primer set was then used to differentiate the three species in a single reaction.
Results: The T-ARMS-PCR assay was able to detect DNA from both species being analysed at the maximum and minimum levels in the DNA ratios (95/5) tested. The duplex T-ARMS-PCR assay was also able to detect all hybrids tested and was validated by sequencing the ITS and the 18S amplicons of 148 of the field samples included in the study.
Conclusions: The duplex tetra-primer ARMS-PCR assay described here can be applied to differentiate between Schistosoma species and their hybrid forms that infect humans and animals, thereby providing a method to investigate the epidemiology of these species in endemic areas. The addition of several markers in a single reaction saves considerable time and is of long-standing interest for investigating genetic populations.
Keywords: Genotyping; Hybridization; SNPs; Schistosomiasis; T-ARMS-PCR.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genetic profiles of Schistosoma haematobium parasites from Malian transmission hotspot areas.Parasit Vectors. 2023 Aug 4;16(1):263. doi: 10.1186/s13071-023-05860-8. Parasit Vectors. 2023. PMID: 37542265 Free PMC article.
-
Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes.PLoS Negl Trop Dis. 2013 Apr 4;7(4):e2110. doi: 10.1371/journal.pntd.0002110. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23593513 Free PMC article.
-
Urogenital schistosomiasis and hybridization between Schistosoma haematobium and Schistosoma bovis in adults living in Richard-Toll, Senegal.Parasitology. 2018 Nov;145(13):1723-1726. doi: 10.1017/S0031182018001415. Epub 2018 Sep 6. Parasitology. 2018. PMID: 30185248
-
[Schistosoma species in Senegal with special reference to the biology, epidemiology and pathology of Schistosoma curassoni Brumpt, 1931].Verh K Acad Geneeskd Belg. 1990;52(1):31-68. Verh K Acad Geneeskd Belg. 1990. PMID: 2191509 Review. Dutch.
-
Mapping of schistosome hybrids of the haematobium group in West and Central Africa.J Helminthol. 2024 Sep 18;98:e53. doi: 10.1017/S0022149X24000257. J Helminthol. 2024. PMID: 39291545 Review.
Cited by
-
Genetic profiles of Schistosoma haematobium parasites from Malian transmission hotspot areas.Parasit Vectors. 2023 Aug 4;16(1):263. doi: 10.1186/s13071-023-05860-8. Parasit Vectors. 2023. PMID: 37542265 Free PMC article.
-
Population genetic structure of Schistosoma bovis and S. curassoni collected from cattle in Mali.Parasite. 2024;31:36. doi: 10.1051/parasite/2024035. Epub 2024 Jul 2. Parasite. 2024. PMID: 38953782 Free PMC article.
-
Schistosoma mansoni x S. haematobium hybrids frequently infecting sub-Saharan migrants in southeastern Europe: Egg DNA genotyping assessed by RD-PCR, sequencing and cloning.PLoS Negl Trop Dis. 2025 Mar 31;19(3):e0012942. doi: 10.1371/journal.pntd.0012942. eCollection 2025 Mar. PLoS Negl Trop Dis. 2025. PMID: 40163525 Free PMC article.
-
Molecular detection and identification of Trichobilharzia: development of a LAMP, qPCR, and multiplex PCR toolkit.Parasit Vectors. 2025 May 30;18(1):195. doi: 10.1186/s13071-025-06822-y. Parasit Vectors. 2025. PMID: 40448150 Free PMC article.
-
Imported Schistosomiasis in Southwestern Europe: Wide Variation of Pure and Hybrid Genotypes Infecting Sub-Saharan Migrants.Transbound Emerg Dis. 2025 Apr 18;2025:6614509. doi: 10.1155/tbed/6614509. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40718473 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

