Parkin regulates microglial NLRP3 and represses neurodegeneration in Parkinson's disease
- PMID: 37029500
- PMCID: PMC10265164
- DOI: 10.1111/acel.13834
Parkin regulates microglial NLRP3 and represses neurodegeneration in Parkinson's disease
Abstract
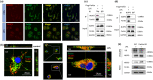
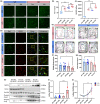
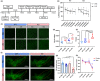
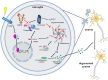
Microglial hyperactivation of the NOD-, LRR-, and pyrin domain-containing 3 (NLRP3) inflammasome contributes to the pathogenesis of Parkinson's disease (PD). Recently, neuronally expressed NLRP3 was demonstrated to be a Parkin polyubiquitination substrate and a driver of neurodegeneration in PD. However, the role of Parkin in NLRP3 inflammasome activation in microglia remains unclear. Thus, we aimed to investigate whether Parkin regulates NLRP3 in microglia. We investigated the role of Parkin in NLRP3 inflammasome activation through the overexpression of Parkin in BV2 microglial cells and knockout of Parkin in primary microglia after lipopolysaccharide (LPS) treatment. Immunoprecipitation experiments were conducted to quantify the ubiquitination levels of NLRP3 under various conditions and to assess the interaction between Parkin and NLRP3. In vivo experiments were conducted by administering intraperitoneal injections of LPS in wild-type and Parkin knockout mice. The Rotarod test, pole test, and open field test were performed to evaluate motor functions. Immunofluorescence was performed for pathological detection of key proteins. Overexpression of Parkin mediated NLRP3 degradation via K48-linked polyubiquitination in microglia. The loss of Parkin activity in LPS-induced mice resulted in excessive microglial NLRP3 inflammasome assembly, facilitating motor impairment, and dopaminergic neuron loss in the substantia nigra. Accelerating Parkin-induced NLRP3 degradation by administration of a heat shock protein (HSP90) inhibitor reduced the inflammatory response. Parkin regulates microglial NLRP3 inflammasome activation through polyubiquitination and alleviates neurodegeneration in PD. These results suggest that targeting Parkin-mediated microglial NLRP3 inflammasome activity could be a potential therapeutic strategy for PD.
Keywords: NLRP3; Parkin; microglia; neuroinflammation; ubiquitination.
© 2023 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
All authors claim that there are no conflicts of interest.
Figures


References
-
- Avagliano, C. , Coretti, L. , Lama, A. , Pirozzi, C. , de Caro, C. , de Biase, D. , Turco, L. , Mollica, M. P. , Paciello, O. , Calignano, A. , Meli, R. , Lembo, F. , & Mattace Raso, G. (2022). Dual‐hit model of Parkinson's disease: Impact of Dysbiosis on 6‐Hydroxydopamine‐insulted mice‐neuroprotective and anti‐inflammatory effects of butyrate. International Journal of Molecular Sciences, 23(12), 6367. 10.3390/ijms23126367 - DOI - PMC - PubMed
-
- Burmann, B. M. , Gerez, J. A. , Matečko‐Burmann, I. , Campioni, S. , Kumari, P. , Ghosh, D. , Mazur, A. , Aspholm, E. E. , Šulskis, D. , Wawrzyniuk, M. , Bock, T. , Schmidt, A. , Rüdiger, S. G. D. , Riek, R. , & Hiller, S. (2020). Regulation of α‐synuclein by chaperones in mammalian cells. Nature, 577(7788), 127–132. 10.1038/s41586-019-1808-9 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

