Hydrophobicity-Driven Increases in Editing in Mitochondrial mRNAs during the Evolution of Kinetoplastids
- PMID: 37030003
- PMCID: PMC10118304
- DOI: 10.1093/molbev/msad081
Hydrophobicity-Driven Increases in Editing in Mitochondrial mRNAs during the Evolution of Kinetoplastids
Abstract
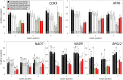
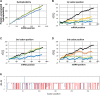
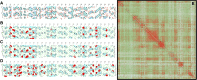
Kinetoplastids are a diverse group of flagellates which exhibit editing by insertion/deletion of Us in the mitochondrial mRNAs. Some mRNAs require editing to build most of their coding sequences, a process known as pan-editing. Evidence suggests that pan-editing is an ancestral feature in kinetoplastids. Here, we investigate how the transition from nonedited to pan-edited states occurred. The mitochondrial mRNAs and protein sequences from nine kinetoplastids and related groups (diplonemids, euglenids, and jakobids) were analyzed. RNA editing increased protein hydrophobicity to extreme values by introducing Us in the second codon position, despite the absence of editing preferences related to codon position. In addition, hydrophobicity was maintained by purifying selection in species that lost editing by retroposition of the fully edited mRNA. Only a few hydrophobic to hydrophilic amino acid changes were inferred for such species. In the protein secondary structure, these changes occurred spatially close to other hydrophilic residues. The analysis of coevolving sites showed that multiple changes are required together for hydrophobicity to be lost, which suggest the proteins are locked into extended hydrophobicity. Finally, an analysis of the NAD7 protein-protein interactions showed they can also influence hydrophobicity increase in the protein and where editing can occur in the mRNA. In conclusion, our results suggest that protein hydrophobicity has influenced editing site selection and how editing expanded in mRNAs. In effect, the hydrophobicity increase was entrenched by a neutral ratchet moved by a mutational pressure to introduce Us, thus helping to explain both RNA editing increase and, possibly, persistence.
Keywords: RNA editing; hydrophobic ratchet; kinetoplastids; pan-editing.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




References
-
- Benne R, van den Burg J, Brakenhoff JP, Sloof P, van Boom JH, Tromp MC. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826. - PubMed
-
- Berger JA, Mitra SK, Carli M, Neri A. 2004. Visualization and analysis of DNA sequences using DNA walks. J Franklin Inst. 341:37–53.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

