Endothelial FAT1 inhibits angiogenesis by controlling YAP/TAZ protein degradation via E3 ligase MIB2
- PMID: 37031213
- PMCID: PMC10082778
- DOI: 10.1038/s41467-023-37671-x
Endothelial FAT1 inhibits angiogenesis by controlling YAP/TAZ protein degradation via E3 ligase MIB2
Abstract
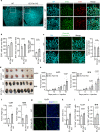
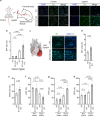
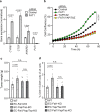
Activation of endothelial YAP/TAZ signaling is crucial for physiological and pathological angiogenesis. The mechanisms of endothelial YAP/TAZ regulation are, however, incompletely understood. Here we report that the protocadherin FAT1 acts as a critical upstream regulator of endothelial YAP/TAZ which limits the activity of these transcriptional cofactors during developmental and tumor angiogenesis by promoting their degradation. We show that loss of endothelial FAT1 results in increased endothelial cell proliferation in vitro and in various angiogenesis models in vivo. This effect is due to perturbed YAP/TAZ protein degradation, leading to increased YAP/TAZ protein levels and expression of canonical YAP/TAZ target genes. We identify the E3 ubiquitin ligase Mind Bomb-2 (MIB2) as a FAT1-interacting protein mediating FAT1-induced YAP/TAZ ubiquitination and degradation. Loss of MIB2 expression in endothelial cells in vitro and in vivo recapitulates the effects of FAT1 depletion and causes decreased YAP/TAZ degradation and increased YAP/TAZ signaling. Our data identify a pivotal mechanism of YAP/TAZ regulation involving FAT1 and its associated E3 ligase MIB2, which is essential for YAP/TAZ-dependent angiogenesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

