RNAi-mediated rheostat for dynamic control of AAV-delivered transgenes
- PMID: 37031257
- PMCID: PMC10082758
- DOI: 10.1038/s41467-023-37774-5
RNAi-mediated rheostat for dynamic control of AAV-delivered transgenes
Abstract
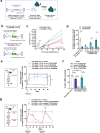
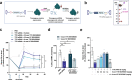
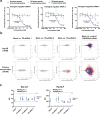
Adeno-associated virus (AAV)-based gene therapy could be facilitated by the development of molecular switches to control the magnitude and timing of expression of therapeutic transgenes. RNA interference (RNAi)-based approaches hold unique potential as a clinically proven modality to pharmacologically regulate AAV gene dosage in a sequence-specific manner. We present a generalizable RNAi-based rheostat wherein hepatocyte-directed AAV transgene expression is silenced using the clinically validated modality of chemically modified small interfering RNA (siRNA) conjugates or vectorized co-expression of short hairpin RNA (shRNA). For transgene induction, we employ REVERSIR technology, a synthetic high-affinity oligonucleotide complementary to the siRNA or shRNA guide strand to reverse RNAi activity and rapidly recover transgene expression. For potential clinical development, we report potent and specific siRNA sequences that may allow selective regulation of transgenes while minimizing unintended off-target effects. Our results establish a conceptual framework for RNAi-based regulatory switches with potential for infrequent dosing in clinical settings to dynamically modulate expression of virally-delivered gene therapies.
© 2023. The Author(s).
Conflict of interest statement
All authors were employees of Alnylam Pharmaceuticals with salary and/or stock options at the time the work was conducted. REVERSIR is a trademark of Alnylam Pharmaceuticals.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

