A novel patient-derived 3D model recapitulates mantle cell lymphoma lymph node signaling, immune profile and in vivo ibrutinib responses
- PMID: 37031299
- PMCID: PMC10244172
- DOI: 10.1038/s41375-023-01885-1
A novel patient-derived 3D model recapitulates mantle cell lymphoma lymph node signaling, immune profile and in vivo ibrutinib responses
Abstract
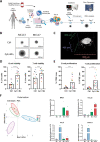
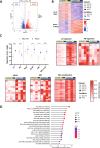
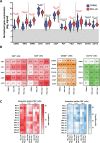
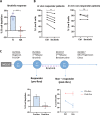
Mantle cell lymphoma (MCL), a rare and aggressive B-cell non-Hodgkin lymphoma, mainly develops in the lymph node (LN) and creates a protective and immunosuppressive niche that facilitates tumor survival, proliferation and chemoresistance. To capture disease heterogeneity and tumor microenvironment (TME) cues, we have developed the first patient-derived MCL spheroids (MCL-PDLS) that recapitulate tumor oncogenic pathways and immune microenvironment in a multiplexed system that allows easy drug screening, including immunotherapies. MCL spheroids, integrated by tumor B cells, monocytes and autologous T-cells self-organize in disc-shaped structures, where B and T-cells maintain viability and proliferate, and monocytes differentiate into M2-like macrophages. RNA-seq analysis demonstrated that tumor cells recapitulate hallmarks of MCL-LN (proliferation, NF-kB and BCR), with T cells exhibiting an exhaustion profile (PD1, TIM-3 and TIGIT). MCL-PDLS reproduces in vivo responses to ibrutinib and demonstrates that combination of ibrutinib with nivolumab (anti-PD1) may be effective in ibrutinib-resistant cases by engaging an immune response with increased interferon gamma and granzyme B release. In conclusion, MCL-PDLS recapitulates specific MCL-LN features and in vivo responses to ibrutinib, representing a robust tool to study MCL interaction with the immune TME and to perform drug screening in a patient-derived system, advancing toward personalized therapeutic approaches.
© 2023. The Author(s).
Conflict of interest statement
MN, RM, PB-L and J-ML are employees of Imactiv3D. The rest of the authors declare no conflict of interest.
Figures


References
-
- Navarro A, Clot G, Royo C, Jares P, Hadzidimitriou A, Agathangelidis A, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72:5307–16. doi: 10.1158/0008-5472.CAN-12-1615. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

